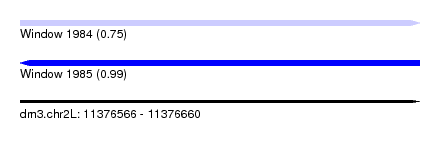
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,376,566 – 11,376,660 |
| Length | 94 |
| Max. P | 0.993003 |

| Location | 11,376,566 – 11,376,660 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.96 |
| Shannon entropy | 0.29024 |
| G+C content | 0.51694 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.81 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
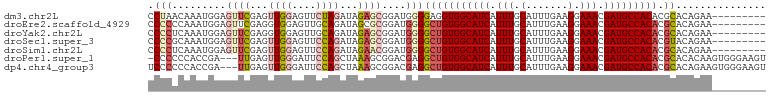
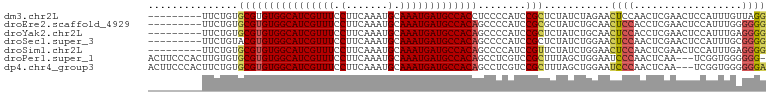
>dm3.chr2L 11376566 94 + 23011544 CCUAACAAAUGGAGUUCGAGUUGGAGUUCUAGAUAGAGCGGAUGGGGAGGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAA--------- ((........)).((((...((((....))))...))))...((.(...((((((((.(((.(.......).))).)))))))).).)).....--------- ( -25.40, z-score = -1.59, R) >droEre2.scaffold_4929 12573877 94 - 26641161 CCCCCCAAAUGGAGUUCGAGGUGGAGUUGCAGAUAGCGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAA--------- .((((((..(.(....).)..)))..((((.......))))..)))(((((((((((.(((.(.......).))).))))))))).))......--------- ( -29.20, z-score = -0.43, R) >droYak2.chr2L 7773497 94 + 22324452 CCCCUCAAAUGGAGUUCGAGGUGGAGUUGCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAA--------- .(((((...((.(..((......))..).))....))).)).((..(((((((((((.(((.(.......).))).))))))))).)).))...--------- ( -28.20, z-score = -0.73, R) >droSec1.super_3 6771022 94 + 7220098 CCCCGCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGUACAGAA--------- ..((((...((((..((......))..))))......)))).((..(((((((((((.(((.(.......).))).))))))))).)).))...--------- ( -31.80, z-score = -2.24, R) >droSim1.chr2L 11182711 94 + 22036055 CCCCUCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAACGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAA--------- (((.((.......((((...((((....))))...)))).)).)))(((((((((((.(((.(.......).))).))))))))).))......--------- ( -33.00, z-score = -2.58, R) >droPer1.super_1 2012490 99 - 10282868 -CCCCCCACCGA---UUGAGUUGGGAUUCCAGCUAAAGCGGACGAGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACACAAGUGGGAAGU -...(((((((.---...((((((....))))))....))).....(((((((((((.(((.(.......).))).))))))))).)).......)))).... ( -37.80, z-score = -3.17, R) >dp4.chr4_group3 4869443 100 - 11692001 UCCCCCCACCGA---UUGAGUUGGGAUUCCAGCUAAAGCGGACGAGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAAGUGGGAAGU ....(((((((.---...((((((....))))))....))).(...(((((((((((.(((.(.......).))).))))))))).))...)...)))).... ( -37.90, z-score = -3.14, R) >consensus CCCCCCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAA_________ .............((((...((((....))))...)))).......(((((((((((.(((.(.......).))).))))))))).))............... (-20.97 = -21.81 + 0.84)
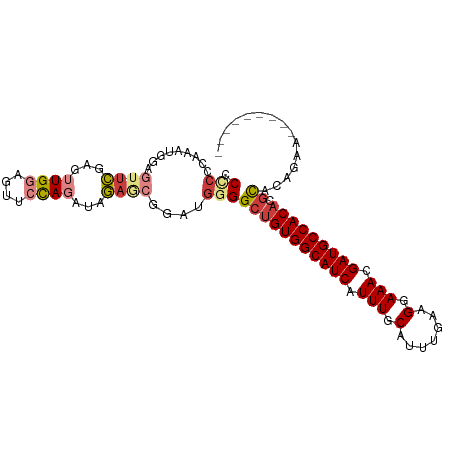
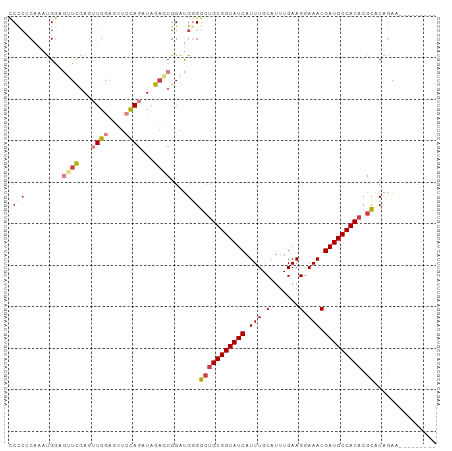
| Location | 11,376,566 – 11,376,660 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.96 |
| Shannon entropy | 0.29024 |
| G+C content | 0.51694 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -19.62 |
| Energy contribution | -21.59 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
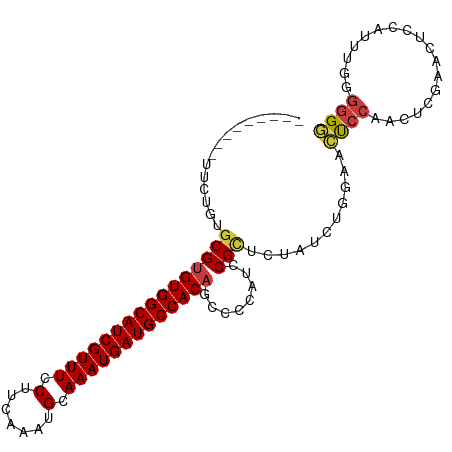
>dm3.chr2L 11376566 94 - 23011544 ---------UUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACCUCCCCAUCCGCUCUAUCUAGAACUCCAACUCGAACUCCAUUUGUUAGG ---------(((((.(((.((((((((((((.(.......).)))))))))))).........)))......)))))..(((((.............))).)) ( -18.22, z-score = -1.67, R) >droEre2.scaffold_4929 12573877 94 + 26641161 ---------UUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCGCUAUCUGCAACUCCACCUCGAACUCCAUUUGGGGGG ---------....((((((((((((((((((.(.......).)))))))))))))........)))))...........((.(((((((.....))))))))) ( -31.90, z-score = -2.79, R) >droYak2.chr2L 7773497 94 - 22324452 ---------UUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGCAACUCCACCUCGAACUCCAUUUGAGGGG ---------......((.(((((((((((((.(.......).))))))))))))))).......((.......))....((.(((((((.....))))))))) ( -31.10, z-score = -3.89, R) >droSec1.super_3 6771022 94 - 7220098 ---------UUCUGUACGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCCAACUCGAACUCCAUUUGCGGGG ---------.........(((((((((((((.(.......).)))))))))))))......(((((......((((..((......))..))))...))))). ( -29.30, z-score = -3.42, R) >droSim1.chr2L 11182711 94 - 22036055 ---------UUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGUUCUAUCUGGAACUCCAACUCGAACUCCAUUUGAGGGG ---------......((.(((((((((((((.(.......).)))))))))))))))(((....(((((....))))).....((((((.....))))))))) ( -33.20, z-score = -4.07, R) >droPer1.super_1 2012490 99 + 10282868 ACUUCCCACUUGUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCUCGUCCGCUUUAGCUGGAAUCCCAACUCAA---UCGGUGGGGGG- .(((((((((((.((((.(((((((((((((.(.......).)))))))))))))))....(((((....)).)))......)).)).---..)))))))))- ( -39.00, z-score = -3.68, R) >dp4.chr4_group3 4869443 100 + 11692001 ACUUCCCACUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCUCGUCCGCUUUAGCUGGAAUCCCAACUCAA---UCGGUGGGGGGA .(((((((((.....((.(((((((((((((.(.......).)))))))))))))))....(((((....)).)))............---..))))))))). ( -39.10, z-score = -3.60, R) >consensus _________UUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCCAACUCGAACUCCAUUUGGGGGG ...............((.(((((((((((((.(.......).)))))))))))))))(((............(((....))).((((((.....))))))))) (-19.62 = -21.59 + 1.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:16 2011