
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,115,435 – 14,115,640 |
| Length | 205 |
| Max. P | 0.790389 |

| Location | 14,115,435 – 14,115,532 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Shannon entropy | 0.14200 |
| G+C content | 0.41332 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14115435 97 + 22422827 CCGCAUCUUCUCUCUUCCGUAGAUAACUGUCCAUGCAUGUGUGUAUGUAUGUCUGCCAUUUCAGCCAGCAUCAUAAAUAUGCAUGUAUGUAUGUACA ..................(((.(((...((.(((((((((...((((.(((.(((.(......).)))))))))).))))))))).)).))).))). ( -23.80, z-score = -1.95, R) >droSim1.chrX 10836426 87 + 17042790 CCGCAUCU---CUCCUCCCUAGAUAACUGUCCAUGCAUGUAUGUAUGUAUGUCUGCCAUUUCAGCCAGCAUCAUAAAUAUGUAUGUUUGU------- ........---....................(((((((((...((((.(((.(((.(......).)))))))))).))))))))).....------- ( -16.00, z-score = -1.15, R) >droSec1.super_20 719476 87 + 1148123 CCGCAUCU---CUCCUCCCUAGAUAACUGUCCAUGCAUGUAUGUAUGUAUGUCUGCCAUUUCAGCCAGCAUCAUAAAUAUGUAUGUAUGU------- ..((((..---..........(((....)))(((((((((...((((.(((.(((.(......).)))))))))).))))))))).))))------- ( -16.40, z-score = -0.98, R) >consensus CCGCAUCU___CUCCUCCCUAGAUAACUGUCCAUGCAUGUAUGUAUGUAUGUCUGCCAUUUCAGCCAGCAUCAUAAAUAUGUAUGUAUGU_______ ..(((................(((....)))(((((((((...((((.(((.(((.(......).)))))))))).)))))))))..)))....... (-17.35 = -17.13 + -0.22)



| Location | 14,115,512 – 14,115,605 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.32 |
| Shannon entropy | 0.51317 |
| G+C content | 0.43948 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14115512 93 + 22422827 -----------------------UAUGCAUGUAUGUAUGUACACACAUAUAGCACCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAGCAGAUAAGCUGCAUAAUAAAC-- -----------------------..(((((((.(((((((....)))))))..........((((((.((....)).))))))...))))))).((((.....)))).........-- ( -26.40, z-score = -1.81, R) >droSim1.chrX 10836500 85 + 17042790 -------------------------------UAUGUAUGUUUGUACAUAUAGCGCCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAGCAGAUAAGCUGCAUAAUAAAC-- -------------------------------..(((((((....)))))))(((.......((((((.((....)).))))))......)))..((((.....)))).........-- ( -20.32, z-score = -0.64, R) >droSec1.super_20 719550 85 + 1148123 -------------------------------UAUGUAUGUAUGUACAUAUAGCGCCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAGCAGAUAAGCUGCAUAAUAAAC-- -------------------------------..((((((((((((((....((((......))))...((....)))))))))...))))))).((((.....)))).........-- ( -21.00, z-score = -0.72, R) >dp4.chrXL_group1e 8461113 118 + 12523060 AAUUCCCACACACCACACAGAUGCACGGACGCAGCCGCAGAGGCAGACACAGCACAGACACAUGCAUGGCUGAGUGUAUGUGUUGCAUAUGCAAGUAGAUAAGCUGCAUAAUAAAGCA .....................(((......((.(((.....)))............(((((((((((......))))))))))))).((((((.((......)))))))).....))) ( -30.70, z-score = 0.15, R) >consensus _______________________________UAUGUAUGUAUGCACAUAUAGCACCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAGCAGAUAAGCUGCAUAAUAAAC__ ..............................((((((((....)))))))).(((.......((((((.((....)).))))))......)))..((((.....))))........... (-16.80 = -16.42 + -0.38)

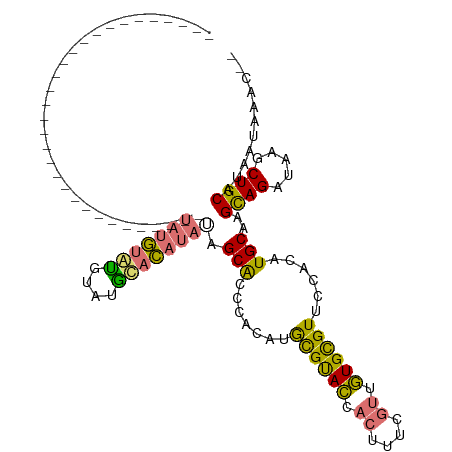
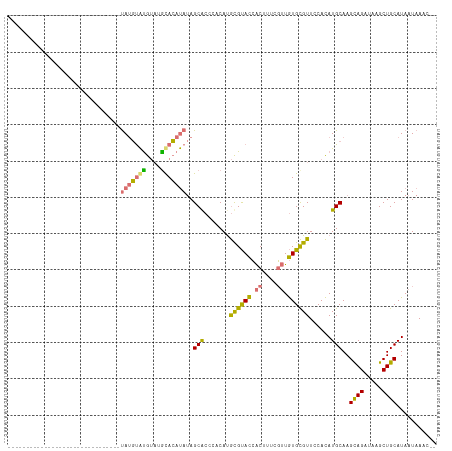
| Location | 14,115,532 – 14,115,640 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Shannon entropy | 0.30273 |
| G+C content | 0.46482 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -22.59 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14115532 108 + 22422827 ---CACAUAUAGCACCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAG-CAGAUAAGCUGCAUAAUAAACCGCA--UUACGAGUGGGUUGCAAAUUGUGAACCUGUG ---((((....((((((((....(.....)..((((.(((((......(((((.(-(......))))))).......))))--).))))))))).)))....))))........ ( -32.72, z-score = -1.92, R) >droSim1.chrX 10836513 107 + 17042790 ----ACAUAUAGCGCCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAG-CAGAUAAGCUGCAUAAUAAACCGCA--UUACGAGUGGAUUGCAAAUUGUGGACCUGUG ----...(((((.(.(((((((((..((((..((((.(((((......(((((.(-(......))))))).......))))--).))))))))..))))...))))).)))))) ( -36.22, z-score = -2.97, R) >droSec1.super_20 719563 107 + 1148123 ----ACAUAUAGCGCCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAG-CAGAUAAGCUGCAUAAUAAACCGCA--UUACGAGUGGGUUGCAAAUUGUGGACCUGUG ----...(((((.(.(((((((((..((((..((((.(((((......(((((.(-(......))))))).......))))--).))))))))..))))...))))).)))))) ( -37.42, z-score = -2.74, R) >droYak2.chrX 8368900 109 + 21770863 ---UACAUAUAUCGCCCACAUGCGUACCACUUCCGAUGUGCGUUCCACAUGCAAGAUAGAUAAGCUGCAUAAUAAACCGCA--UUGCGAGUGGGUUGCAAAUUGUGGACCUGUG ---............(((((((((..((((((.(((((((.(((....(((((............)))))....)))))))--))).))))))..))))...)))))....... ( -34.10, z-score = -2.11, R) >droEre2.scaffold_4690 5933814 108 - 18748788 ---UACAUAUAGCGCCCACAUGCGUACCACUCUCGUUGUGCGUUCCACAUGCAAG-UAGAUAAGCUGCAUAAUAAGCCGCA--UGACGAGUGGGUUGCAAAUUGUGGACCAGUG ---............(((((((((.(((.(.(((((..((((......(((((.(-(......))))))).......))))--..))))).))))))))...)))))....... ( -36.72, z-score = -2.10, R) >dp4.chrXL_group1e 8461153 111 + 12523060 AGGCAGACACAGCACAGACACAUGCAUGGCUGAGUGUAUGUGUUGCAUAUGCAAG-UAGAUAAGCUGCAUAAUAAAGCACACAUUACGAGUAGGUUG--AGUUGUGUUGUUGUU .(((((.((((((.(((((((((((((......)))))))))))...((((((.(-(......))))))))........................))--.)))))))))))... ( -32.40, z-score = -0.29, R) >consensus ____ACAUAUAGCGCCCACAUGCGUACCACUUUCGUUGUGCGUUCCACAUGCAAG_CAGAUAAGCUGCAUAAUAAACCGCA__UUACGAGUGGGUUGCAAAUUGUGGACCUGUG .......(((((...(((((((((..((((((......((((......(((((............))))).......))))......))))))..))))...)))))..))))) (-22.59 = -22.82 + 0.23)
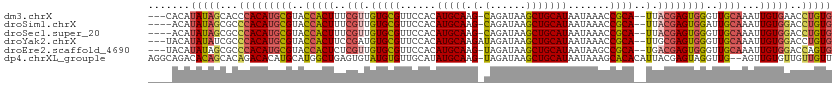


| Location | 14,115,532 – 14,115,640 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Shannon entropy | 0.30273 |
| G+C content | 0.46482 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -24.65 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14115532 108 - 22422827 CACAGGUUCACAAUUUGCAACCCACUCGUAA--UGCGGUUUAUUAUGCAGCUUAUCUG-CUUGCAUGUGGAACGCACAACGAAAGUGGUACGCAUGUGGGUGCUAUAUGUG--- ........((((....(((.(((((.((((.--((((.(((((.(((((((......)-).)))))))))).))))..((....))..))))...))))))))....))))--- ( -36.60, z-score = -1.93, R) >droSim1.chrX 10836513 107 - 17042790 CACAGGUCCACAAUUUGCAAUCCACUCGUAA--UGCGGUUUAUUAUGCAGCUUAUCUG-CUUGCAUGUGGAACGCACAACGAAAGUGGUACGCAUGUGGGCGCUAUAUGU---- ...(((((((((...(((...((((((((..--((((.(((((.(((((((......)-).)))))))))).))))..))))..))))...)))))))))).))......---- ( -39.80, z-score = -3.09, R) >droSec1.super_20 719563 107 - 1148123 CACAGGUCCACAAUUUGCAACCCACUCGUAA--UGCGGUUUAUUAUGCAGCUUAUCUG-CUUGCAUGUGGAACGCACAACGAAAGUGGUACGCAUGUGGGCGCUAUAUGU---- ...(((((((((...(((...((((((((..--((((.(((((.(((((((......)-).)))))))))).))))..))))..))))...)))))))))).))......---- ( -39.10, z-score = -2.63, R) >droYak2.chrX 8368900 109 - 21770863 CACAGGUCCACAAUUUGCAACCCACUCGCAA--UGCGGUUUAUUAUGCAGCUUAUCUAUCUUGCAUGUGGAACGCACAUCGGAAGUGGUACGCAUGUGGGCGAUAUAUGUA--- .....(((((((...(((...(((((((...--((((.(((((.((((((..........))))))))))).))))...))..)))))...))))))))))..........--- ( -32.00, z-score = -0.68, R) >droEre2.scaffold_4690 5933814 108 + 18748788 CACUGGUCCACAAUUUGCAACCCACUCGUCA--UGCGGCUUAUUAUGCAGCUUAUCUA-CUUGCAUGUGGAACGCACAACGAGAGUGGUACGCAUGUGGGCGCUAUAUGUA--- .....(((((((...(((.((((.(((((..--((((((.......))......((((-(......))))).))))..))))).).)))..))))))))))..........--- ( -36.50, z-score = -1.72, R) >dp4.chrXL_group1e 8461153 111 - 12523060 AACAACAACACAACU--CAACCUACUCGUAAUGUGUGCUUUAUUAUGCAGCUUAUCUA-CUUGCAUAUGCAACACAUACACUCAGCCAUGCAUGUGUCUGUGCUGUGUCUGCCU ...............--..........(((.(((((((.....(((((((........-.))))))).)).))))))))((.((((((.((....)).)).)))).))...... ( -22.80, z-score = 0.07, R) >consensus CACAGGUCCACAAUUUGCAACCCACUCGUAA__UGCGGUUUAUUAUGCAGCUUAUCUA_CUUGCAUGUGGAACGCACAACGAAAGUGGUACGCAUGUGGGCGCUAUAUGU____ .....(((((((...(((...(((((.......((((.(((((.((((((..........))))))))))).)))).......)))))...))))))))))............. (-24.65 = -25.57 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:26 2011