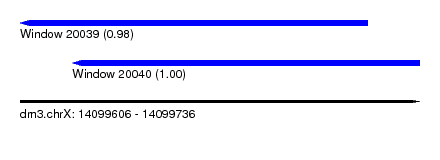
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,099,606 – 14,099,736 |
| Length | 130 |
| Max. P | 0.999406 |

| Location | 14,099,606 – 14,099,719 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.83 |
| Shannon entropy | 0.20794 |
| G+C content | 0.50557 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -33.12 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14099606 113 - 22422827 AGAUUUUCCGGCUUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUAUCGUGGUGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUG------GCGAGUGAAAUGCGAGAUUGGC- ((((..(((((...(((..(((((..(((((((((..((((..........))))..))))))))))))))...)))..)))))..)))).------((.(((..........))).))- ( -40.80, z-score = -2.69, R) >droEre2.scaffold_4690 5917818 111 + 18748788 AGAUUUUCCGGCAUCAAGGGCCCAAGAACGUUGGGUCUGCCCGCAUGUCGG--UGGGCUCAAUGUCUGGGUAAAUUG-ACUGGAUUAUCUG------GCGAGUGAAAUGCGAGAUUGGCC ((((..(((((..((((..(((((.((.(((((((((..((........))--..))))))))))))))))...)))-))))))..))))(------((.(((..........))).))) ( -45.40, z-score = -2.94, R) >droYak2.chrX 8353305 118 - 21770863 AGAUUUUCCGGCUUCAAGGGCCCAGGAACGUUGAGUUUGCCAGCAUGUUGG--UGGGCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUGUGAAAUGCGAGUGAAAUGCGAGAUGGGCA ((((..(((((...(((..((((((..((((((((((..((((....))))--..))))))))))))))))...)))..)))))..)))).................(((.......))) ( -46.40, z-score = -3.85, R) >droSec1.super_20 704102 113 - 1148123 AGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGCUGGUCUCAAUGUCUGGGUAAAUUGUACUGGACUAUCUG------GCGAGUGAAAUGCGAGAUUGGC- ......(((((...(((..(((((..(((((((((...(((.(((.....))).)))))))))))))))))...)))..)))))(.((((.------(((.......))).)))).)..- ( -42.80, z-score = -2.59, R) >droSim1.chrX_random 3771578 113 - 5698898 AGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGUUGCUCUCAAUGCCUGGGUAAAUUGUACUGGACUAUCUG------GCGAGUGAAAUGCGAGAUUGGC- ......(((((...(((..(((((.((.(((((((..((((((.....)))).))..))))))))))))))...)))..)))))(.((((.------(((.......))).)))).)..- ( -39.60, z-score = -1.70, R) >consensus AGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGG_UGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUG______GCGAGUGAAAUGCGAGAUUGGC_ ((((..(((((...(((..((((.(((.(((((((..((((((.....)))).))..))))))))))))))...)))..)))))..)))).......(((.......))).......... (-33.12 = -32.20 + -0.92)


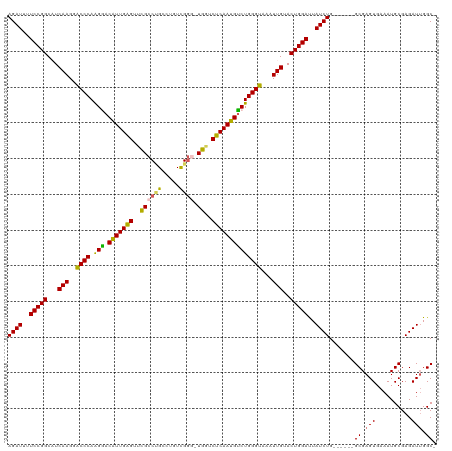
| Location | 14,099,623 – 14,099,736 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Shannon entropy | 0.24071 |
| G+C content | 0.48344 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -36.94 |
| Energy contribution | -36.42 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.66 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14099623 113 - 22422827 CUCAA-UCAAUUGCAGUUAGAUUUUCCGGCUUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUAUCGUGGUGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUGGCGAG------ .....-.........(((((((..(((((...(((..(((((..(((((((((..((((..........))))..))))))))))))))...)))..)))))..)))))))...------ ( -45.90, z-score = -4.98, R) >droEre2.scaffold_4690 5917836 110 + 18748788 AUCAA-UCAAUUUCAGUUAGAUUUUCCGGCAUCAAGGGCCCAAGAACGUUGGGUCUGCCCGCAUGUCGG--UGGGCUCAAUGUCUGGGUAAAUUG-ACUGGAUUAUCUGGCGAG------ .....-.........(((((((..(((((..((((..(((((.((.(((((((((..((........))--..))))))))))))))))...)))-))))))..)))))))...------ ( -47.20, z-score = -4.89, R) >droYak2.chrX 8353323 118 - 21770863 CUCAACCCAAUUUCAGUUAGAUUUUCCGGCUUCAAGGGCCCAGGAACGUUGAGUUUGCCAGCAUGUUGG--UGGGCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUGUGAAAUGCGAG (((......((((((..(((((..(((((...(((..((((((..((((((((((..((((....))))--..))))))))))))))))...)))..)))))..)))))))))))..))) ( -50.40, z-score = -5.45, R) >droSec1.super_20 704119 113 - 1148123 CUCAA-UCAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGCUGGUCUCAAUGUCUGGGUAAAUUGUACUGGACUAUCUGGCGAG------ .....-.........(((((((..(((((...(((..(((((..(((((((((...(((.(((.....))).)))))))))))))))))...)))..)))))..)))))))...------ ( -46.50, z-score = -4.41, R) >droSim1.chrX_random 3771595 113 - 5698898 CUCAA-UCAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGGUUGCUCUCAAUGCCUGGGUAAAUUGUACUGGACUAUCUGGCGAG------ .....-.........(((((((..(((((...(((..(((((.((.(((((((..((((((.....)))).))..))))))))))))))...)))..)))))..)))))))...------ ( -43.30, z-score = -3.58, R) >consensus CUCAA_UCAAUUGCAGUUAGAUUUUCCGGCAUCAAGGACCCAAGGACAUUGAGUUCGCCUGCCUGUCGGG_UGGUCUCAAUGUCUGGGUAAAUUGUACUGGAUUAUCUGGCGAG______ ...............(((((((..(((((...(((..((((.(((.(((((((..((((((.....)))).))..))))))))))))))...)))..)))))..)))))))......... (-36.94 = -36.42 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:20 2011