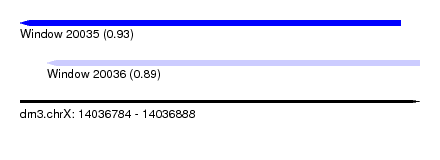
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,036,784 – 14,036,888 |
| Length | 104 |
| Max. P | 0.925977 |

| Location | 14,036,784 – 14,036,883 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.17 |
| Shannon entropy | 0.61874 |
| G+C content | 0.49380 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
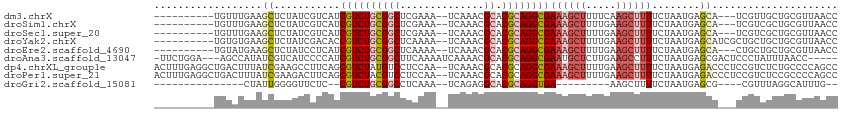
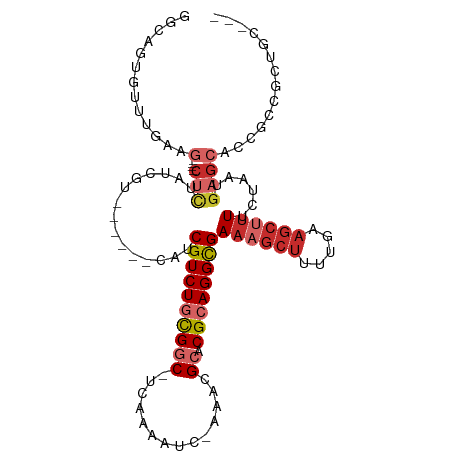
>dm3.chrX 14036784 99 - 22422827 ----------UGUUUGAAGCUCUAUCGUCAUCGUCUGCGGCUCGAAA--UCAAACGCACGCAGGCGAAAGCUUUUCAAGCUUUUCUAAUGAGCA---UCGUUGCUGCGUUAACC ----------.((((((((((.........(((((((((((......--......)).))))))))).))))..)))))).....(((((((((---....)))).)))))... ( -30.10, z-score = -1.65, R) >droSim1.chrX 10786371 99 - 17042790 ----------UGUUUGAAGCUCUAUCGUCAUCGUCUGCGGCUCGAAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCA---UCGUCGCUGCGUUAACC ----------.....(((((((....((..(((((((((((......--......)).)))))))))..))....).))))))..((((((((.---.....))).)))))... ( -28.80, z-score = -0.78, R) >droSec1.super_20 647710 99 - 1148123 ----------UGUUUGAAGCUCUAUCGUCAUCGUCUGCGGCUCGAAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCA---UCGUCGCUGCGUUAACC ----------.....(((((((....((..(((((((((((......--......)).)))))))))..))....).))))))..((((((((.---.....))).)))))... ( -28.80, z-score = -0.78, R) >droYak2.chrX 8296121 102 - 21770863 ----------UGUGUGAAGCUCUAUCGACACCGUCUGCGGCUCAAAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCAUCGCUGCUGCUGCGUUAACC ----------...((((.((((.........((((((((((......--......)).))))))))((((((.....))))))......)))).)))).((....))....... ( -31.90, z-score = -1.26, R) >droEre2.scaffold_4690 5857713 99 + 18748788 ----------UGUAUGAAGCUCUAUCCUCAUCGUCUGCGGCUCAAAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCA---CUGCUGCUGCGUUAACC ----------........((((........(((((((((((......--......)).)))))))))(((((.....))))).......)))).---..((....))....... ( -28.00, z-score = -1.34, R) >droAna3.scaffold_13047 1550226 105 - 1816235 -UUCUGGA---AGCCAUAUCGUCAUCCCCAUCGUCUGCGGCUUCAAAAUCAAAACGCACGCAGGCGAAUGCUCUUGAAGCCUUUCUAAUGAGCGACUCCCUAUUUAACC----- -.......---.........(((.......(((((((((((..............)).)))))))))..((((..(((....)))....))))))).............----- ( -25.84, z-score = -1.82, R) >dp4.chrXL_group1e 6324890 112 + 12523060 ACUUUGAGGCUGACUUUAUCGAAGCCUUCAGCGUCUAUGUCCUCCAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGACCCUCCGUCUCUGCCCCAGCC .......(((((...(((..(((((.((((((((..((........)--)...))))....((((....)))).)))))))))..))).(((((.....))))).....))))) ( -29.50, z-score = -1.45, R) >droPer1.super_21 479655 112 + 1645244 ACUUUGAGGCUGACUUUAUCGAAGACUUCAGCGUCUACGUCCUCCAA--UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGACCCUCCGUCUCCGCCCCAGCC .......(((((........((((((((((((((...(((.......--....))).))))((((....)))).))))).)))))....(((((.....))))).....))))) ( -31.90, z-score = -2.29, R) >droGri2.scaffold_15081 2905114 80 + 4274704 ---------------CUAUUGGGGUUCUC--CGUCUGCGGCCUCAAA--UCAGAGGCACGCAGGUGA---------AAGCUUUUCUAAUGAGCG----CGUUUAGGCAUUUG-- ---------------.((((((((..((.--(..((((((((((...--...))))).)))))..).---------.))...))))))))...(----(......)).....-- ( -26.00, z-score = -1.47, R) >consensus __________UGUCUGAAGCUCUAUCGUCAUCGUCUGCGGCUCCAAA__UCAAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCA___CCGUCGCUGCGUUAACC ..................((...........((((((((((..............)).))))))))((((((.....))))))........))..................... (-13.96 = -14.75 + 0.79)
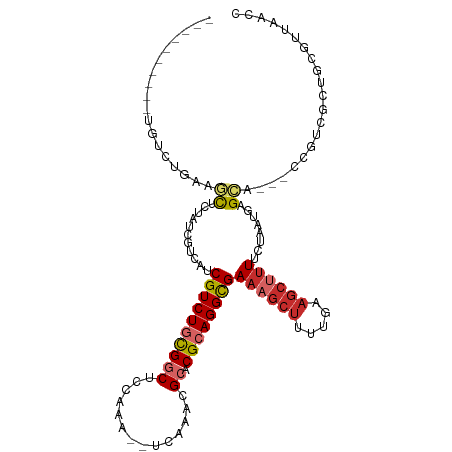

| Location | 14,036,791 – 14,036,888 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.53 |
| Shannon entropy | 0.58089 |
| G+C content | 0.50446 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
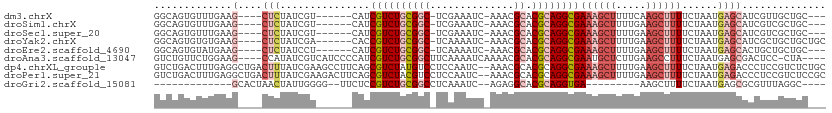
>dm3.chrX 14036791 97 - 22422827 GGCAGUGUUUGAAG----CUCUAUCGU------CAUCGUCUGCGGC-UCGAAAUC-AAACGCACGCAGGCGAAAGCUUUUCAAGCUUUUCUAAUGAGCAUCGUUGCUGC--- .((((((..(((.(----(((......------..(((((((((((-........-....)).)))))))))(((((.....))))).......)))).))).))))))--- ( -34.40, z-score = -2.37, R) >droSim1.chrX 10786378 97 - 17042790 GGCAGUGUUUGAAG----CUCUAUCGU------CAUCGUCUGCGGC-UCGAAAUC-AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCAUCGUCGCUGC--- .((((((..(((.(----(((......------..(((((((((((-........-....)).)))))))))(((((.....))))).......)))).))).))))))--- ( -36.80, z-score = -2.72, R) >droSec1.super_20 647717 97 - 1148123 GGCAGUGUUUGAAG----CUCUAUCGU------CAUCGUCUGCGGC-UCGAAAUC-AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCAUCGUCGCUGC--- .((((((..(((.(----(((......------..(((((((((((-........-....)).)))))))))(((((.....))))).......)))).))).))))))--- ( -36.80, z-score = -2.72, R) >droYak2.chrX 8296128 100 - 21770863 GGCAGUGUGUGAAG----CUCUAUCGA------CACCGUCUGCGGC-UCAAAAUC-AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCAUCGCUGCUGCUGC (((((((.((((.(----(((......------...((((((((((-........-....)).))))))))((((((.....))))))......)))).))))))))))).. ( -39.60, z-score = -2.82, R) >droEre2.scaffold_4690 5857720 97 + 18748788 GGCAGUGUAUGAAG----CUCUAUCCU------CAUCGUCUGCGGC-UCAAAAUC-AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCACUGCUGCUGC--- ((((((((..((((----(((......------..(((((((((((-........-....)).)))))))))........).))))))((....)))))))))).....--- ( -33.49, z-score = -2.16, R) >droAna3.scaffold_13047 1550233 103 - 1816235 GUCUGUUCUGGAAG----CCAUAUCGUCAUCCCCAUCGUCUGCGGCUUCAAAAUCAAAACGCACGCAGGCGAAUGCUCUUGAAGCCUUUCUAAUGAGCGACUCC-CUA---- (((.((((.(((((----.................(((((((((((..............)).)))))))))..(((.....))))))))....)))))))...-...---- ( -27.64, z-score = -1.86, R) >dp4.chrXL_group1e 6324897 110 + 12523060 GUCUGACUUUGAGGCUGACUUUAUCGAAGCCUUCAGCGUCUAUGUCCUCCAAUC--AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGACCCUCCGUCUCUGC (((((....(((((((...........)))).)))((((..((........)).--..))))...)))))(((((((.....))))))).....(((((.....)))))... ( -27.50, z-score = -0.58, R) >droPer1.super_21 479662 110 + 1645244 GUCUGACUUUGAGGCUGACUUUAUCGAAGACUUCAGCGUCUACGUCCUCCAAUC--AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGACCCUCCGUCUCCGC ....(((...((((....(((....((((((((((((((...(((.........--..))).))))((((....)))).))))).)))))....)))..)))).)))..... ( -26.30, z-score = -0.16, R) >droGri2.scaffold_15081 2905119 82 + 4274704 -------------GCACUAACUAUUGGGG--UUCUCCGUCUGCGGCCUCAAAUC--AGAGGCACGCAGGUGA---------AAGCUUUUCUAAUGAGCGCGUUUAGGC---- -------------((.((.......((((--((...(..((((((((((.....--.))))).)))))..).---------.)))))).......)).))........---- ( -27.54, z-score = -1.61, R) >consensus GGCAGUGUUUGAAG____CUCUAUCGU______CAUCGUCUGCGGC_UCAAAAUC_AAACGCACGCAGGCGAAAGCUUUUGAAGCUUUUCUAAUGAGCACCGCCGCUGC___ .((.................................((((((((...................))))))))((((((.....))))))........)).............. (-12.96 = -13.74 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:17 2011