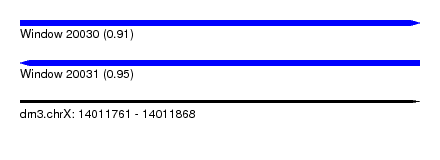
| Sequence ID | dm3.chrX |
|---|---|
| Location | 14,011,761 – 14,011,868 |
| Length | 107 |
| Max. P | 0.950966 |

| Location | 14,011,761 – 14,011,868 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.37766 |
| G+C content | 0.55918 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.99 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14011761 107 + 22422827 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GAUGGGUGGGUGGCGAAAAUGAG ..((.((((((((.......)))))........((((..((((((.....))))))..))))..(((((((.((((((....))----).))).))))))))))...)).. ( -35.30, z-score = -0.52, R) >droSim1.chrX 10759750 107 + 17042790 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GAUGGGUGCGUGCCGAAAAUGAA ..((.((((((((.......)))((((.((((.((((..((((((.....))))))..))))....(((((....)))..))..----..)))).)))))))))...)).. ( -37.00, z-score = -1.47, R) >droSec1.super_20 622777 107 + 1148123 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GAUGGUUGCGUGCCGAAAAUGAA .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..----..((((.....))))........ ( -35.02, z-score = -1.21, R) >droEre2.scaffold_4690 5833230 107 - 18748788 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GACGAGAGGGUGGCGAAAAUAGC .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..----..(....)............... ( -32.42, z-score = 0.05, R) >droYak2.chrX 8270535 107 + 21770863 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GAUGAGAGGGUGGCGAAAAUAGG .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..----....................... ( -32.02, z-score = 0.14, R) >droAna3.scaffold_13047 1514254 107 + 1816235 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----CUUGUGGGUCUUGCGACUAAAGA .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).(----(((...((((....)))).)))) ( -37.72, z-score = -1.70, R) >dp4.chrXL_group1e 6294573 105 - 12523060 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCCU----UAUGUCCAUGAUCUUUAUAAA-- .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..----.....................-- ( -31.62, z-score = -1.93, R) >droPer1.super_21 429333 90 - 1645244 CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU----GAUGGG----------------- .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..----......----------------- ( -32.02, z-score = -1.79, R) >droWil1.scaffold_181096 3900415 111 - 12416693 CGCAGCUGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCGCUCGGCCACAACUGCUUCUGUGAACUGCGGGUCGUCGGAAUGGG ...(((..(((((.......))))))))((((.((((..((((((.....))))))..))))..(((..(((((.....(((....(....).)))))))).)))..)))) ( -34.90, z-score = 0.30, R) >droVir3.scaffold_12970 10259196 83 - 11907090 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCU---------------------------- .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))).---------------------------- ( -27.02, z-score = -1.60, R) >droGri2.scaffold_15081 1434960 83 + 4274704 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGGCUGCU---------------------------- .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))))))).---------------------------- ( -34.12, z-score = -2.96, R) >anoGam1.chr3L 33610019 94 + 41284009 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUU----GGUGGAGGAG------------- ........(((((.......))))).((((...((((..((((((.....))))))..))))..(((((.(((.......))).----))))).))))------------- ( -31.70, z-score = -0.80, R) >consensus CGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUU____GAUGGG_GGGUGGCGAAAAU___ .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))............................. (-31.00 = -30.99 + -0.01)

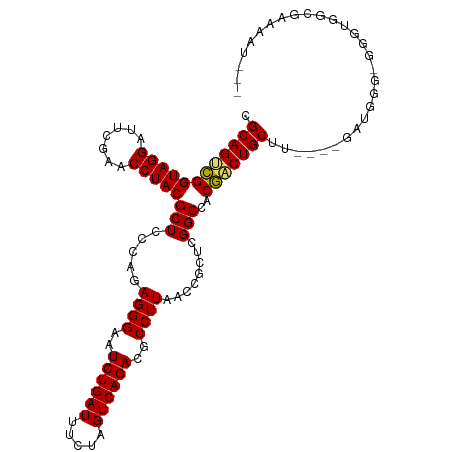
| Location | 14,011,761 – 14,011,868 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.37766 |
| G+C content | 0.55918 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -33.11 |
| Energy contribution | -32.66 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 14011761 107 - 22422827 CUCAUUUUCGCCACCCACCCAUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .......................----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.90, z-score = -1.02, R) >droSim1.chrX 10759750 107 - 17042790 UUCAUUUUCGGCACGCACCCAUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .......(((((.(((((.....----.....)).))))))))((((((...((((((((.......))))))((((...))))))(((((.......))))).)))))). ( -34.90, z-score = -1.10, R) >droSec1.super_20 622777 107 - 1148123 UUCAUUUUCGGCACGCAACCAUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .......(((((.(((.((....----.....)).))))))))((((((...((((((((.......))))))((((...))))))(((((.......))))).)))))). ( -35.60, z-score = -1.44, R) >droEre2.scaffold_4690 5833230 107 + 18748788 GCUAUUUUCGCCACCCUCUCGUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .......................----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.90, z-score = -0.39, R) >droYak2.chrX 8270535 107 - 21770863 CCUAUUUUCGCCACCCUCUCAUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .......................----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.90, z-score = -0.89, R) >droAna3.scaffold_13047 1514254 107 - 1816235 UCUUUAGUCGCAAGACCCACAAG----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG ((((..(((....)))....)))----).((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -39.10, z-score = -2.28, R) >dp4.chrXL_group1e 6294573 105 + 12523060 --UUUAUAAAGAUCAUGGACAUA----AGGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG --.....................----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.70, z-score = -1.06, R) >droPer1.super_21 429333 90 + 1645244 -----------------CCCAUC----AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG -----------------......----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.90, z-score = -2.03, R) >droWil1.scaffold_181096 3900415 111 + 12416693 CCCAUUCCGACGACCCGCAGUUCACAGAAGCAGUUGUGGCCGAGCGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCAGCUGCG ((((..........((((.(..(((((......)))))..)..))))....((.((((((.......)))))).))...))))((((((((.......)))))..)))... ( -33.40, z-score = 0.01, R) >droVir3.scaffold_12970 10259196 83 + 11907090 ----------------------------AGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ----------------------------.((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.54, R) >droGri2.scaffold_15081 1434960 83 - 4274704 ----------------------------AGCAGCCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ----------------------------.((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -35.00, z-score = -2.27, R) >anoGam1.chr3L 33610019 94 - 41284009 -------------CUCCUCCACC----AAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG -------------..........----..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -0.70, R) >consensus ___AUUUUCGCCACCC_CCCAUC____AAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCG .............................((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). (-33.11 = -32.66 + -0.45)
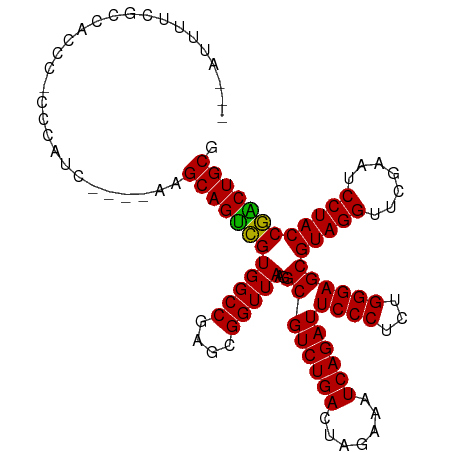
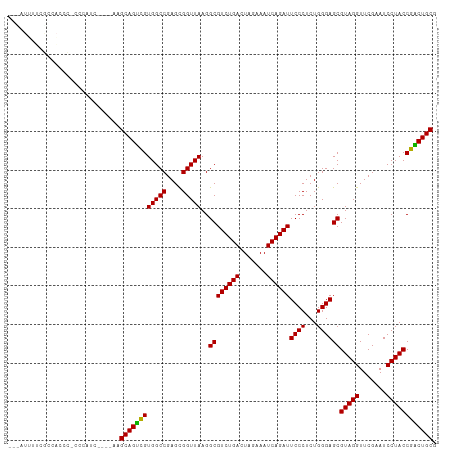
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:13 2011