
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,998,680 – 13,998,763 |
| Length | 83 |
| Max. P | 0.788236 |

| Location | 13,998,680 – 13,998,763 |
|---|---|
| Length | 83 |
| Sequences | 11 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 93.49 |
| Shannon entropy | 0.15060 |
| G+C content | 0.56968 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -27.68 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
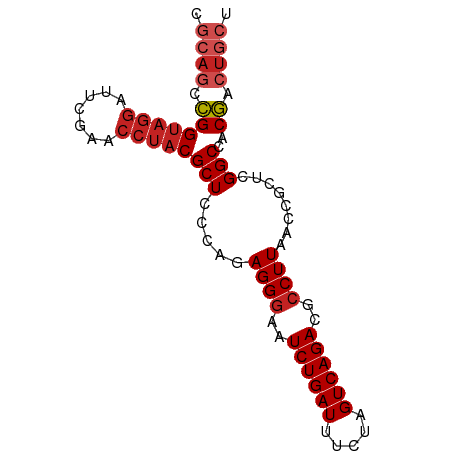
>dm3.chrX 13998680 83 + 22422827 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.30, z-score = -1.71, R) >droSec1.super_20 617706 76 + 1148123 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUACCUCUGGGAGCG-------GGAGCCUACCGGCUGCG .((((((((((((....)))))((((..(((.((.....(((((.....))))).))))-------)..))))..))))))). ( -31.80, z-score = -1.83, R) >droYak2.chrX 8266661 83 + 21770863 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.30, z-score = -1.71, R) >droEre2.scaffold_4690 5829054 83 - 18748788 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.30, z-score = -1.71, R) >droAna3.scaffold_13047 1509218 83 + 1816235 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -32.30, z-score = -1.71, R) >dp4.chrXL_group1e 6288586 77 - 12523060 AACAGUCGUGGCCGAGAGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCG------ ....(((.(((((....))))).)))((((((.......))))))((((...))))(.(((((.......)))))).------ ( -24.90, z-score = -1.12, R) >droPer1.super_21 423796 83 - 1645244 AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGAUUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -30.60, z-score = -1.78, R) >droWil1.scaffold_180699 2076170 83 - 2593675 AGCAGUUGUGGCCGAGUGGUUAAGGCGUCUGACUUGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCAGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -30.20, z-score = -1.90, R) >droMoj3.scaffold_6496 15765523 83 + 26866924 AGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.54, R) >droGri2.scaffold_15245 13573130 80 + 18325388 AGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCU--- .........(((((..........((((((((.......))))))((((...))))))(((((.......))))))))))--- ( -24.00, z-score = 0.32, R) >anoGam1.chr3L 33597567 83 + 41284009 AGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.54, R) >consensus AGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). (-27.68 = -28.45 + 0.78)



| Location | 13,998,680 – 13,998,763 |
|---|---|
| Length | 83 |
| Sequences | 11 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 93.49 |
| Shannon entropy | 0.15060 |
| G+C content | 0.56968 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13998680 83 - 22422827 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.36, R) >droSec1.super_20 617706 76 - 1148123 CGCAGCCGGUAGGCUCC-------CGCUCCCAGAGGUAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.((...((((..-------.((.....(((((..((((((.....)))))).)))))....))..)))).)).)))). ( -25.90, z-score = -1.45, R) >droYak2.chrX 8266661 83 - 21770863 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.36, R) >droEre2.scaffold_4690 5829054 83 + 18748788 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.36, R) >droAna3.scaffold_13047 1509218 83 - 1816235 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.36, R) >dp4.chrXL_group1e 6288586 77 + 12523060 ------CGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCUCUCGGCCACGACUGUU ------..(((((.......))))).....(((((((..((((((.....))))))..))))......(((....)))))).. ( -21.10, z-score = -0.53, R) >droPer1.super_21 423796 83 + 1645244 CGCAAUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .(((.((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..))).))). ( -26.02, z-score = -1.55, R) >droWil1.scaffold_180699 2076170 83 + 2593675 CGCAGCUGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCAAGUCAGACGCCUUAACCACUCGGCCACAACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -26.02, z-score = -2.07, R) >droMoj3.scaffold_6496 15765523 83 - 26866924 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.60, R) >droGri2.scaffold_15245 13573130 80 - 18325388 ---AGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCU ---.(((((((((.......)))))........((((..((((((.....))))))..)))).......)))).......... ( -24.20, z-score = -1.09, R) >anoGam1.chr3L 33597567 83 - 41284009 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). ( -27.02, z-score = -1.60, R) >consensus CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCU .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).)))). (-22.66 = -23.58 + 0.93)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:11 2011