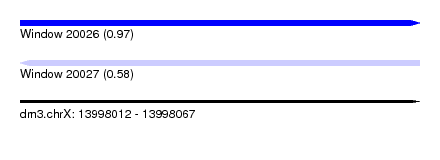
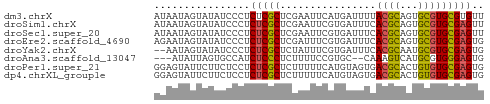
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,998,012 – 13,998,067 |
| Length | 55 |
| Max. P | 0.966164 |

| Location | 13,998,012 – 13,998,067 |
|---|---|
| Length | 55 |
| Sequences | 8 |
| Columns | 55 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.49923 |
| G+C content | 0.46910 |
| Mean single sequence MFE | -12.44 |
| Consensus MFE | -8.09 |
| Energy contribution | -7.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
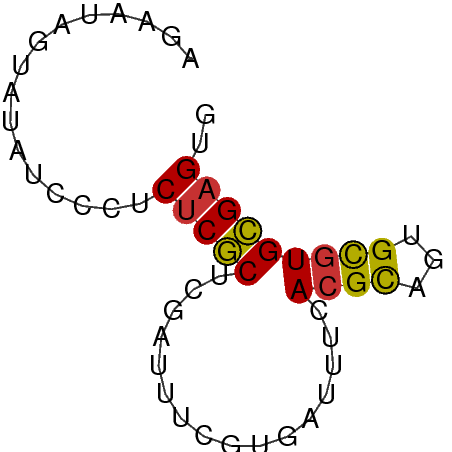
>dm3.chrX 13998012 55 + 22422827 AACACGCACGCACUGCGUAAAAUCAUGAAUUCGAGCGAGAGGGAUAUACUAUUAU ...(((((.....)))))...(((.....(((......))).))).......... ( -6.50, z-score = 0.51, R) >droSim1.chrX 10753784 55 + 17042790 AACUCGCACGCACUGCGUGAAAUCACGAAUUCGAGCGAGAGGGAUAUACUAUUAU ..(((((.((.....((((....))))....)).)))))................ ( -15.30, z-score = -2.51, R) >droSec1.super_20 617199 55 + 1148123 AACUCGCACGCACUGCGUGAAAUCACGAAUUCGAGCGAGAGGGAUAUACUAUUAU ..(((((.((.....((((....))))....)).)))))................ ( -15.30, z-score = -2.51, R) >droEre2.scaffold_4690 5828440 55 - 18748788 CACUCGCACGCACUGCGUGAAAUCACGAAAUCGAGCGAGAGGGAUAUACUAUUCU ..(((((.((.....((((....))))....)).))))).((((((...)))))) ( -16.20, z-score = -2.57, R) >droYak2.chrX 8266009 53 + 21770863 CACUCGCACGCAUUGCGUGAAAUCACGAAAUAGAGCGAGAGGGAUAUACUAUU-- ..(((((.(..(((.((((....)))).))).).)))))..............-- ( -14.20, z-score = -2.80, R) >droAna3.scaffold_13047 1508632 50 + 1816235 CACUCCCACGCAUGACUUUG--GCACGGAAAAGAGGGAGAUGGCACUAAUAU--- (((((((..((.........--)).(......).))))).))..........--- ( -9.00, z-score = -0.55, R) >droPer1.super_21 423083 55 - 1645244 CACUCGCACACAGUGCGUCACUACAUGAAAAAGAGCGAGAGGAGAAGAAUACUCC ..(((((....((((...))))...(....)...))))).((((.......)))) ( -11.50, z-score = -1.28, R) >dp4.chrXL_group1e 6287862 55 - 12523060 CACUCGCACACAGUGCGUCACUACAUGAAAAAGAGCGAGAGGAGAAGAAUACUCC ..(((((....((((...))))...(....)...))))).((((.......)))) ( -11.50, z-score = -1.28, R) >consensus CACUCGCACGCACUGCGUGAAAUCACGAAAUAGAGCGAGAGGGAUAUACUAUUAU ..(((((.(......(((......))).....).)))))................ ( -8.09 = -7.94 + -0.16)
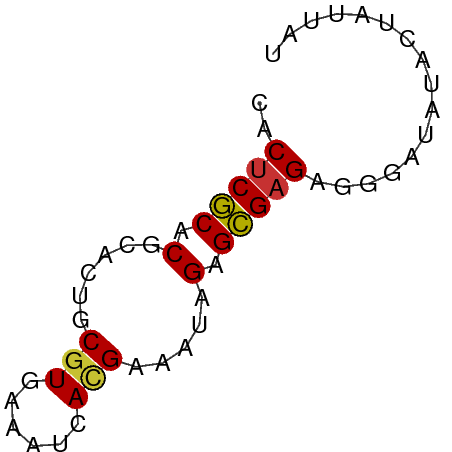
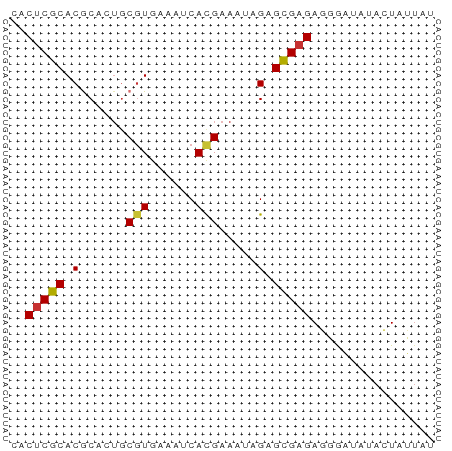
| Location | 13,998,012 – 13,998,067 |
|---|---|
| Length | 55 |
| Sequences | 8 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Shannon entropy | 0.49923 |
| G+C content | 0.46910 |
| Mean single sequence MFE | -13.50 |
| Consensus MFE | -5.97 |
| Energy contribution | -5.70 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13998012 55 - 22422827 AUAAUAGUAUAUCCCUCUCGCUCGAAUUCAUGAUUUUACGCAGUGCGUGCGUGUU ....................................((((((.....)))))).. ( -6.00, z-score = 1.18, R) >droSim1.chrX 10753784 55 - 17042790 AUAAUAGUAUAUCCCUCUCGCUCGAAUUCGUGAUUUCACGCAGUGCGUGCGAGUU ................(((((.((.(((((((....)))).))).)).))))).. ( -16.40, z-score = -2.70, R) >droSec1.super_20 617199 55 - 1148123 AUAAUAGUAUAUCCCUCUCGCUCGAAUUCGUGAUUUCACGCAGUGCGUGCGAGUU ................(((((.((.(((((((....)))).))).)).))))).. ( -16.40, z-score = -2.70, R) >droEre2.scaffold_4690 5828440 55 + 18748788 AGAAUAGUAUAUCCCUCUCGCUCGAUUUCGUGAUUUCACGCAGUGCGUGCGAGUG ................(((((.(((((.((((....)))).))).)).))))).. ( -16.40, z-score = -2.09, R) >droYak2.chrX 8266009 53 - 21770863 --AAUAGUAUAUCCCUCUCGCUCUAUUUCGUGAUUUCACGCAAUGCGUGCGAGUG --..............(((((.(((((.((((....)))).)))).).))))).. ( -14.70, z-score = -3.04, R) >droAna3.scaffold_13047 1508632 50 - 1816235 ---AUAUUAGUGCCAUCUCCCUCUUUUCCGUGC--CAAAGUCAUGCGUGGGAGUG ---..........((.(((((.(.....(((((--....).)))).).))))))) ( -8.10, z-score = -0.08, R) >droPer1.super_21 423083 55 + 1645244 GGAGUAUUCUUCUCCUCUCGCUCUUUUUCAUGUAGUGACGCACUGUGUGCGAGUG ((((.......))))...(((((.....((..(((((...)))))..)).))))) ( -15.00, z-score = -1.81, R) >dp4.chrXL_group1e 6287862 55 + 12523060 GGAGUAUUCUUCUCCUCUCGCUCUUUUUCAUGUAGUGACGCACUGUGUGCGAGUG ((((.......))))...(((((.....((..(((((...)))))..)).))))) ( -15.00, z-score = -1.81, R) >consensus AGAAUAGUAUAUCCCUCUCGCUCGAUUUCGUGAUUUCACGCAGUGCGUGCGAGUG ................(((((................((((...))))))))).. ( -5.97 = -5.70 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:10 2011