
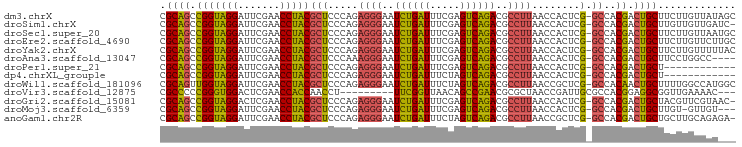
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,997,395 – 13,997,490 |
| Length | 95 |
| Max. P | 0.942589 |

| Location | 13,997,395 – 13,997,490 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Shannon entropy | 0.39937 |
| G+C content | 0.56503 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -25.51 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
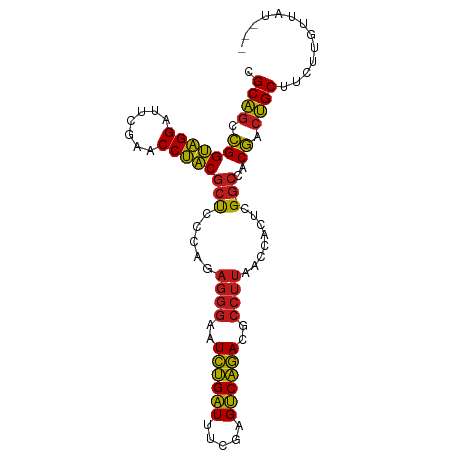
>dm3.chrX 13997395 95 + 22422827 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUCUUGUUAUAGC .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).))))............. ( -27.02, z-score = -0.71, R) >droSim1.chrX 10752861 94 + 17042790 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUGUUGUUGAUC- ....(((((((((.......)))))........((((..((((((.....))))))..)))).......))-)).(((((.....))))).....- ( -27.60, z-score = -0.58, R) >droSec1.super_20 616608 95 + 1148123 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUCUUGUUAAUGC .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).))))............. ( -27.02, z-score = -0.75, R) >droEre2.scaffold_4690 5827856 95 - 18748788 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUCUUGUUCUUGC .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).))))............. ( -27.02, z-score = -0.47, R) >droYak2.chrX 8265415 95 + 21770863 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUCUUGUUUUUAC .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).))))............. ( -27.02, z-score = -1.14, R) >droAna3.scaffold_13047 1507846 91 + 1816235 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAAAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUCCUGGCC---- ((..(((((((((.......)))))........((((..((((((.....))))))..)))).......))-))..))...(((....))).---- ( -27.10, z-score = -0.82, R) >droPer1.super_21 421577 83 - 1645244 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCU------------ .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).)))).------------ ( -27.02, z-score = -1.60, R) >dp4.chrXL_group1e 6286405 83 - 12523060 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCACUCG-GCCACGACUGCU------------ .((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..)).)))).------------ ( -27.02, z-score = -1.91, R) >droWil1.scaffold_181096 3899200 95 - 12416693 CGCAGUUGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCG-GCCACAACUGCUUUUGGCCAUGGC .((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)-))..))))))).....((....)) ( -30.92, z-score = -1.46, R) >droVir3.scaffold_12875 15833734 84 - 20611582 CGCCCCCGGGUGGACUCGAACCACCAACCU---------UUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCGGUUGAAAAC--- ((((.((((((((.......))))).....---------((((((.....)))))).((((.........))))..))).)))).........--- ( -33.00, z-score = -2.36, R) >droGri2.scaffold_15081 2874486 94 - 4274704 CGCAGCCGGUAGGACUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUACGUUCGUAAC- .((((.((((..((((((((.......((((...)))).......))))))))..))(((..........)-))..)).))))(((....)))..- ( -29.04, z-score = -1.18, R) >droMoj3.scaffold_6359 2936747 91 + 4525533 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG-GCCACGACUGCUUGU-GUUGU--- ....(((((((((.......)))))........((((..((((((.....))))))..)))).......))-)).(((((.......-)))))--- ( -27.20, z-score = -0.43, R) >anoGam1.chr2R 51361699 94 - 62725911 CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCG-GCCACGACUGCUGCUUGCAGAGA- ((..(((((((((.......)))))((......((((..((((((.....))))))..))))....)).))-))..)).((((.....))))...- ( -32.50, z-score = -1.41, R) >consensus CGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCG_GCCACGACUGCUUCUUGUUAU___ .((((.(((((((.......)))))........((((..((((((.....))))))..))))..............)).))))............. (-25.51 = -24.90 + -0.61)
| Location | 13,997,395 – 13,997,490 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Shannon entropy | 0.39937 |
| G+C content | 0.56503 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -28.23 |
| Energy contribution | -27.29 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
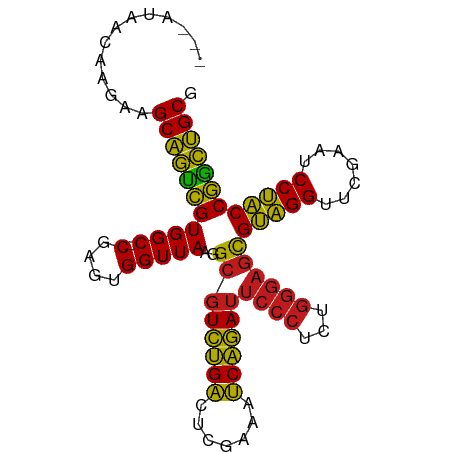
>dm3.chrX 13997395 95 - 22422827 GCUAUAACAAGAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .............(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -0.97, R) >droSim1.chrX 10752861 94 - 17042790 -GAUCAACAACAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG -............(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.13, R) >droSec1.super_20 616608 95 - 1148123 GCAUUAACAAGAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .............(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.02, R) >droEre2.scaffold_4690 5827856 95 + 18748788 GCAAGAACAAGAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .............(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -0.80, R) >droYak2.chrX 8265415 95 - 21770863 GUAAAAACAAGAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .............(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.43, R) >droAna3.scaffold_13047 1507846 91 - 1816235 ----GGCCAGGAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUUUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ----(((((.((.....)).))))-)..(..(((...((((((((.......))))))((((...))))))(((((.......))))).)))..). ( -33.40, z-score = -1.02, R) >droPer1.super_21 421577 83 + 1645244 ------------AGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ------------.(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.54, R) >dp4.chrXL_group1e 6286405 83 + 12523060 ------------AGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ------------.(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.78, R) >droWil1.scaffold_181096 3899200 95 + 12416693 GCCAUGGCCAAAAGCAGUUGUGGC-CGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCAACUGCG ((((..((........))..))))-...((((((...((((((((.......))))))((((...))))))(((((.......))))).)))))). ( -34.10, z-score = -2.00, R) >droVir3.scaffold_12875 15833734 84 + 20611582 ---GUUUUCAACCGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAA---------AGGUUGGUGGUUCGAGUCCACCCGGGGGCG ---.........((((((((.(((((.........)))).((((((..((((((...---------.))))))..).)))))....).)))))))) ( -37.90, z-score = -2.53, R) >droGri2.scaffold_15081 2874486 94 + 4274704 -GUUACGAACGUAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAGUCCUACCGGCUGCG -(((((....)))))...((..((-((.((((...((....))((((((((....(.(((((...)))))).....))))))))))))))))..)) ( -36.80, z-score = -1.94, R) >droMoj3.scaffold_6359 2936747 91 - 4525533 ---ACAAC-ACAAGCAGUCGUGGC-CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG ---.....-....(((((((((((-(....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))). ( -31.40, z-score = -1.22, R) >anoGam1.chr2R 51361699 94 + 62725911 -UCUCUGCAAGCAGCAGUCGUGGC-CGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG -...((((.....)))).((..((-((.((..((((((.((((((.......))))))..))).)))..))(((((.......)))))))))..)) ( -33.50, z-score = -0.83, R) >consensus ___AUAACAAGAAGCAGUCGUGGC_CGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCG .............(((((((.................((((((((.......))))))((((...))))))(((((.......)))))))))))). (-28.23 = -27.29 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:08 2011