| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,991,269 – 13,991,361 |
| Length | 92 |
| Max. P | 0.818840 |

| Location | 13,991,269 – 13,991,361 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.52 |
| Shannon entropy | 0.18489 |
| G+C content | 0.50332 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.16 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13991269 92 - 22422827 AUUGCGCAUACGCACUGUGCGCCCUGUGCACCCUGCAGCUUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUC ...(((((..((((..((((((...))))))..))).)..))))).-..(((((.........)))))....((((((......))))))... ( -29.80, z-score = -1.53, R) >droEre2.scaffold_4690 5822031 92 + 18748788 AUUGCGCAUACGCACUGUGCGCCCUGUGCACCCUGCAAGUUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUC .(((((((((.(((..((((((...))))))..)))..((((((..-...)))))).)))))..))))....((((((......))))))... ( -30.80, z-score = -1.99, R) >droYak2.chrX 8258078 92 - 21770863 AUUGCGCAUACGCACUGUGCGCCCUGUGCACCCUGCAGCUUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUC ...(((((..((((..((((((...))))))..))).)..))))).-..(((((.........)))))....((((((......))))))... ( -29.80, z-score = -1.53, R) >droSec1.super_20 610812 92 - 1148123 AUUGCGCAUACGCACUGUGCGCCCUGUGCACCCUGCAGCUUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUC ...(((((..((((..((((((...))))))..))).)..))))).-..(((((.........)))))....((((((......))))))... ( -29.80, z-score = -1.53, R) >droWil1.scaffold_181096 8583237 78 - 12416693 ---GCGCAUACGCACUGUGCACUCU-----------AGAAUGCGCCUUUUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUUAAA-ACUGACAUU ---((((((..((.....)).(...-----------.).))))))(..((((((.........))))))..)((((((.....-))))))... ( -20.90, z-score = -1.33, R) >droPer1.super_21 414901 89 + 1645244 AUUGCGCAUACGCACUGUGCACCCU-UGCACCCUGCAGCAUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAA--ACUGACAUC ...((((((.((((..(((((....-)))))..))).).)))))).-..(((((.........)))))....((((((....--))))))... ( -29.10, z-score = -2.17, R) >dp4.chrXL_group1e 6279812 89 + 12523060 AUUGCGCAUACGCACUGUGCACCCU-UGCACCCUGCAGCAUGCGCC-UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAA--ACUGACAUC ...((((((.((((..(((((....-)))))..))).).)))))).-..(((((.........)))))....((((((....--))))))... ( -29.10, z-score = -2.17, R) >consensus AUUGCGCAUACGCACUGUGCGCCCUGUGCACCCUGCAGCUUGCGCC_UAUUGCAAUAUAUGCCUGCAGAUUGGUCAGUUCAAACACUGACAUC ...(((((...(((..(((((.....)))))..)))....)))))....(((((.........)))))....((((((......))))))... (-24.18 = -25.16 + 0.98)

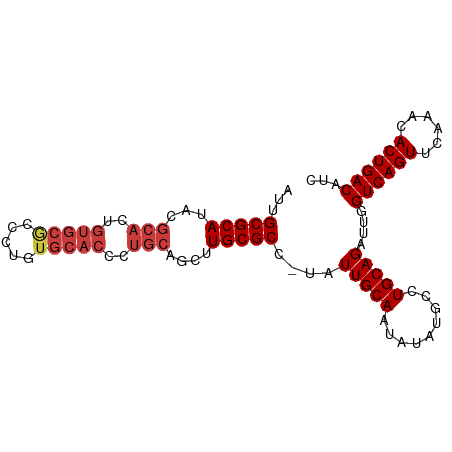
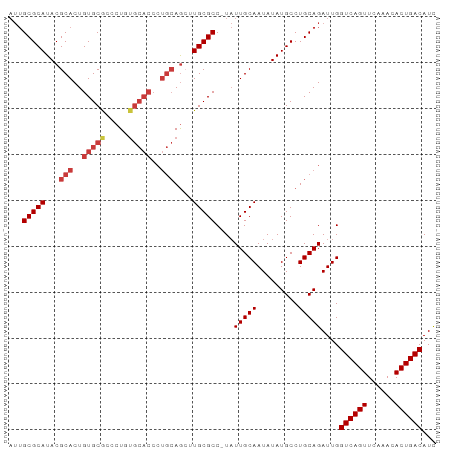
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:06 2011