| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,989,086 – 13,989,190 |
| Length | 104 |
| Max. P | 0.959522 |

| Location | 13,989,086 – 13,989,190 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.27 |
| Shannon entropy | 0.25747 |
| G+C content | 0.44429 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
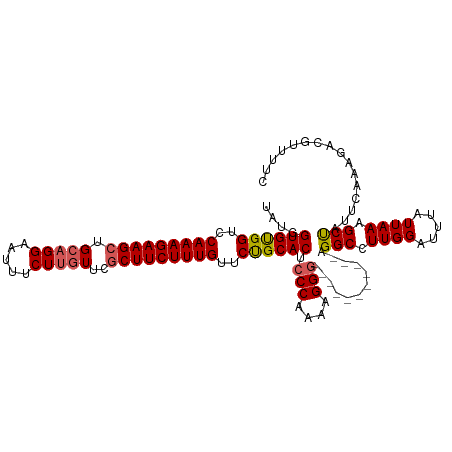
>dm3.chrX 13989086 104 + 22422827 UAUGGUGUGGUCCAAAGAAGCUGCAGGAGUUUCUUAUUUUCUUCUUUGUUCUGCACUCCC-AAAGGG-----------AGGCCUUGGAUUUAUUAAAGCUAUUCAAAGACAUUUUC ...((((..(..((((((((....(((.....))).....))))))))..)..).(((((-...)))-----------)))))((((((...........)))))).......... ( -25.50, z-score = -0.39, R) >droSim1.chrU 1737924 105 - 15797150 UAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCCCCAAAGGG-----------AGGCCUUGGAUUUAUUAAAGCCAUUCAAAGACGUUUUG .((((((..(..(((((((((.(((((.....)))))..)))))))))..)..).(((((....)))-----------)).................))))).............. ( -35.90, z-score = -3.01, R) >droSec1.super_20 608694 105 + 1148123 UAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCCCCAAAGGC-----------AGGCCUUGGAUUUAUUAAAGCCAUUCUAAGACGUUUUG .((((((..(..(((((((((.(((((.....)))))..)))))))))..)..).....((((.((.-----------...))))))..........))))).............. ( -33.70, z-score = -2.65, R) >droYak2.chrX 8255830 112 + 21770863 ----GUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCCCAAAGGGGGCGAAAUGGCGAGGCCUUGGAUUUAUUAAAGCUAUUCAAAGACGUUUUC ----(((..(..(((((((((.(((((.....)))))..)))))))))..)..))).......(..((((....(((...)))((((((...........))))))...))))..) ( -36.50, z-score = -2.00, R) >droEre2.scaffold_4690 5819984 115 - 18748788 UACGGUGCGGUCCAAAGAAGCUGCAGGAAUUUCUGGUUCGCUUCUUUGUUCCGCACUCCCAAAGGGGACGACCCGGUGAGGCCUUGGAUAUAUUAAAGCUAUUCAAAGACGUUUG- .(((((((((..(((((((((.((.((.....)).))..)))))))))..)))))).((((..(((.....)))..)).))..(((((((.........)))))))...)))...- ( -40.30, z-score = -2.35, R) >consensus UAUGGUGUGGUCCAAAGAAGCUGCAGGAAUUUCUUGUUCGCUUCUUUGUUCUGCACUCCCAAAAGGG___________AGGCCUUGGAUUUAUUAAAGCUAUUCAAAGACGUUUUC ...(((((((..(((((((((.(((((.....)))))..)))))))))..)))))))(((....)))............(((.((((.....)))).)))................ (-28.92 = -29.36 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:05 2011