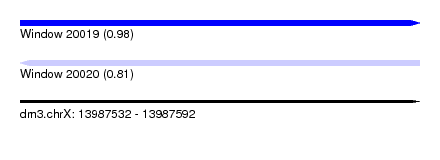
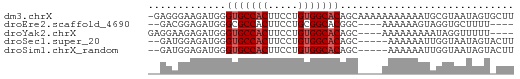
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,987,532 – 13,987,592 |
| Length | 60 |
| Max. P | 0.979212 |

| Location | 13,987,532 – 13,987,592 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 73.08 |
| Shannon entropy | 0.45369 |
| G+C content | 0.46830 |
| Mean single sequence MFE | -12.26 |
| Consensus MFE | -12.22 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
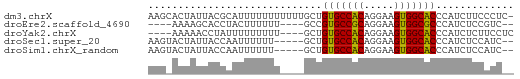
>dm3.chrX 13987532 60 + 22422827 AAGCACUAUUACGCAUUUUUUUUUUUGCUGUGCCACAGGAAGUGGCACCCAUCUUCCCUC- ............(((..........))).(((((((.....)))))))............- ( -13.60, z-score = -1.97, R) >droEre2.scaffold_4690 5818490 51 - 18748788 ----AAAAGCACCUACUUUUUU----GCCGUGCCGCAGGAAGUGGCGCCCAUCUCCGUC-- ----....(((..........)----)).(((((((.....)))))))...........-- ( -11.70, z-score = -0.23, R) >droYak2.chrX 8253856 53 + 21770863 ----AAAAACCUAUUUUUUUUU----GCUGUGCCACAGGAAGUGGCACCCAUCUCUUCCUC ----..................----...(((((((.....)))))))............. ( -12.00, z-score = -2.08, R) >droSec1.super_20 607221 54 + 1148123 AAGUACUAUUACCAAUUUUUU-----GCUGUGCCACAGGAAGUGGCACCCAUCUCCAUC-- .....................-----...(((((((.....)))))))...........-- ( -12.00, z-score = -1.52, R) >droSim1.chrX_random 3751563 54 + 5698898 AAGUACUAUUACCAAUUUUUU-----GCUGUGCCACAGGAAGUGGCACCCAUCUCCAUC-- .....................-----...(((((((.....)))))))...........-- ( -12.00, z-score = -1.52, R) >consensus AAG_ACUAUUACCAAUUUUUUU____GCUGUGCCACAGGAAGUGGCACCCAUCUCCAUC__ .............................(((((((.....)))))))............. (-12.22 = -11.90 + -0.32)
| Location | 13,987,532 – 13,987,592 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 73.08 |
| Shannon entropy | 0.45369 |
| G+C content | 0.46830 |
| Mean single sequence MFE | -14.04 |
| Consensus MFE | -11.28 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
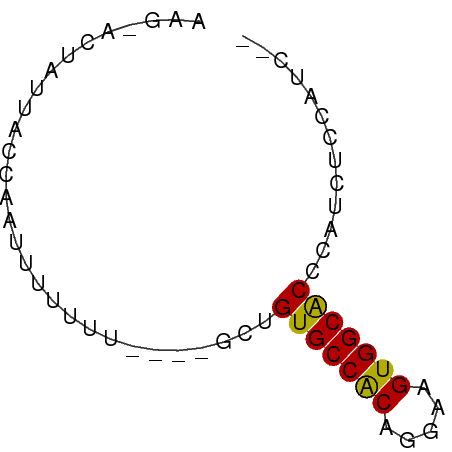
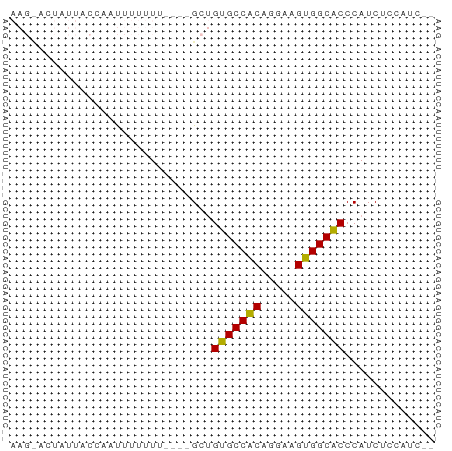
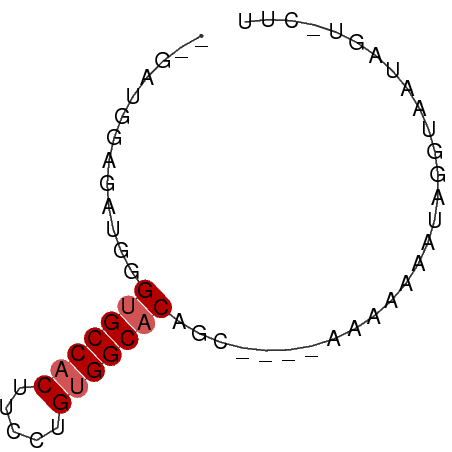
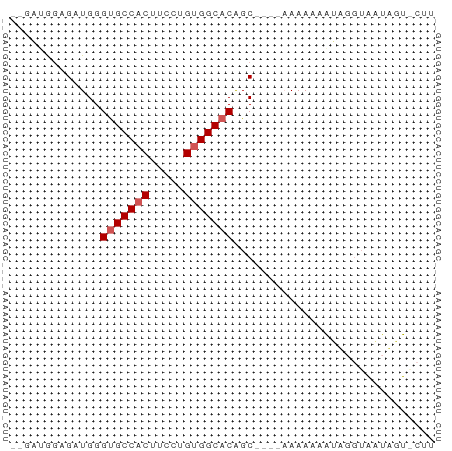
>dm3.chrX 13987532 60 - 22422827 -GAGGGAAGAUGGGUGCCACUUCCUGUGGCACAGCAAAAAAAAAAAUGCGUAAUAGUGCUU -..((((((.(((...)))))))))..(((((.(((..........)))......))))). ( -14.70, z-score = -0.95, R) >droEre2.scaffold_4690 5818490 51 + 18748788 --GACGGAGAUGGGCGCCACUUCCUGCGGCACGGC----AAAAAAGUAGGUGCUUUU---- --.........(((((((((((..(((......))----)...)))).)))))))..---- ( -14.50, z-score = -0.22, R) >droYak2.chrX 8253856 53 - 21770863 GAGGAAGAGAUGGGUGCCACUUCCUGUGGCACAGC----AAAAAAAAAUAGGUUUUU---- ..........((.(((((((.....)))))))..)----).................---- ( -13.60, z-score = -1.37, R) >droSec1.super_20 607221 54 - 1148123 --GAUGGAGAUGGGUGCCACUUCCUGUGGCACAGC-----AAAAAAUUGGUAAUAGUACUU --........((.(((((((.....)))))))..)-----).......((((....)))). ( -13.70, z-score = -0.98, R) >droSim1.chrX_random 3751563 54 - 5698898 --GAUGGAGAUGGGUGCCACUUCCUGUGGCACAGC-----AAAAAAUUGGUAAUAGUACUU --........((.(((((((.....)))))))..)-----).......((((....)))). ( -13.70, z-score = -0.98, R) >consensus __GAUGGAGAUGGGUGCCACUUCCUGUGGCACAGC____AAAAAAAUAGGUAAUAGU_CUU .............(((((((.....)))))))............................. (-11.28 = -11.68 + 0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:04 2011