
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,986,098 – 13,986,191 |
| Length | 93 |
| Max. P | 0.826612 |

| Location | 13,986,098 – 13,986,191 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.05 |
| Shannon entropy | 0.50302 |
| G+C content | 0.39544 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -12.86 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13986098 93 + 22422827 ---------CUUCAUUCACCCAACUCCAGUGCUUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGC-GAAU----- ---------....((((.....((....))(((((((((...(((......))).((((((((((((......)))))))))))).)))))...))))-))))----- ( -19.30, z-score = -0.73, R) >droEre2.scaffold_4690 5817140 96 - 18748788 -----------CUCGCCAGCCAACUGCAGUGCUUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAACUGAGUGAAU- -----------.(((((((....)))((((...((((((...(((......))).((((((((((((......)))))))))))).))))))....)))).))))..- ( -23.70, z-score = -1.02, R) >droSec1.super_20 605783 93 + 1148123 ---------CUCCAUCCGCCCAACUCCAGUGCUUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGC-GAAU----- ---------.......(((.((....(((((((((....)))))...........((((((((((((......))))))))))))))))....)).))-)...----- ( -19.50, z-score = -0.52, R) >droSim1.chrX_random 3749131 93 + 5698898 ---------CUCCAUCCGCCCAACUCCAGUGCUUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGC-GAAU----- ---------.......(((.((....(((((((((....)))))...........((((((((((((......))))))))))))))))....)).))-)...----- ( -19.50, z-score = -0.52, R) >droPer1.super_21 406848 102 - 1645244 CUUCCUCUCCCUGUCCUCUUUUGGGCAGGCUCCUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGCAAAA------ ..........((((((......))))))((((.((((((...(((......))).((((((((((((......)))))))))))).))))))..))))....------ ( -31.80, z-score = -2.84, R) >dp4.chrXL_group1e 6272890 102 - 12523060 CUUCCUCUCCCUGUCCUCUUUUGGGCAGGCUCCUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGCAAAA------ ..........((((((......))))))((((.((((((...(((......))).((((((((((((......)))))))))))).))))))..))))....------ ( -31.80, z-score = -2.84, R) >droVir3.scaffold_12970 10246433 87 - 11907090 ---------------CUCAGGCCGAGUGGCUAUUUUAGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGCAAAAAA------ ---------------..((((((....)))..((((((....(((......))).((((((((((((......)))))))))))).))))))))).......------ ( -19.70, z-score = -1.16, R) >droMoj3.scaffold_6359 1664790 87 - 4525533 ---------------CUUCUUGCAGUUGGCAGUUUUAGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGCAAAAAA------ ---------------....((((((((..(((((((......(((......)))....(((((((((......)))))))))))))))).))))))))....------ ( -25.90, z-score = -3.30, R) >droGri2.scaffold_15081 2862844 97 - 4274704 ----------UCCAGUCCAUUCACCAUACU-CUUUUAGGCUAGCAUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGCAAAAAGCGUGCA ----------................(((.-(((((..((..((........)).((((((((((((......)))))))))))).........)).))))).))).. ( -16.70, z-score = 0.31, R) >consensus _________CUCCAUCCCCCCAACUCCAGUGCUUUUCGGAAAGCGUUUAAUCGCAUUUACAUAGAUUAGGCAGAAUUUGUGUGAAAUUGAAACUGAGCAAAA______ ..........................................(((......))).((((((((((((......))))))))))))....................... (-12.86 = -12.97 + 0.11)
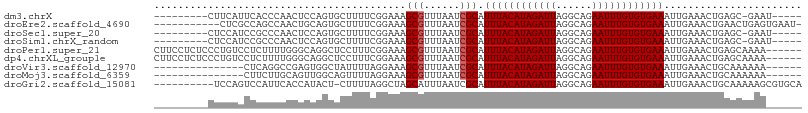

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:03 2011