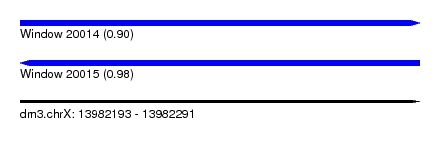
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,982,193 – 13,982,291 |
| Length | 98 |
| Max. P | 0.976163 |

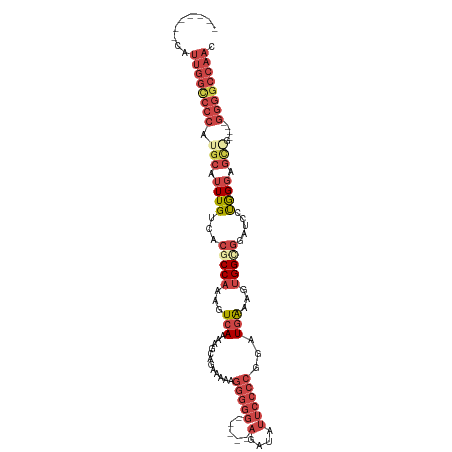
| Location | 13,982,193 – 13,982,291 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.46086 |
| G+C content | 0.56120 |
| Mean single sequence MFE | -34.29 |
| Consensus MFE | -15.84 |
| Energy contribution | -18.50 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
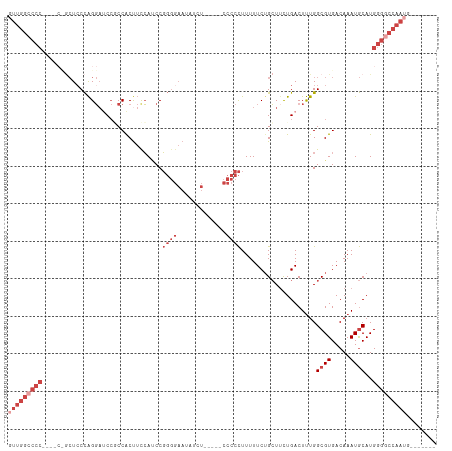
>dm3.chrX 13982193 98 + 22422827 GUUGGCCCC----ACGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUG------- (((((((((----(.((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....)).)))))))))).------- ( -38.10, z-score = -3.35, R) >droSim1.chrX_random 3745339 98 + 5698898 GUUGGCCCC----CUGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUG------- (((((((((----.(((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....))).))))))))).------- ( -38.10, z-score = -3.33, R) >droSec1.super_20 601901 98 + 1148123 GUUGGCCCC----CUGCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU-----CCCCCUUUUUCUGCUUUUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUG------- (((((((((----.(((((....))(((((((.........(((((.....)-----)))).....((.......))...))))).)).....))).))))))))).------- ( -38.10, z-score = -3.33, R) >droYak2.chrX 8248527 107 + 21770863 UUUGGCCCCAAACCCGCUGCCAGGAACCUCCACUUUCAACCGGGGAAUAUAUAUCUCCCCCCUUUUUCUGCUGCUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUG------- .(((((((((.....((.((..((.....))..........(((((.........))))).........)).))........((((.....)))).)))))))))..------- ( -35.00, z-score = -2.40, R) >droEre2.scaffold_4690 5812938 97 - 18748788 GUUGGCCCC----C-ACUCCCAGGCUCCACCACUUUCAUCCGGGGAAUAUGU-----CCCCCUUUUUCAGCUUCUGACUUUGGCGUGACAAAUGCAUGGGACCAAUG------- (((((....----.-..(((((.((.....(((........(((((.....)-----))))........(((.........))))))......)).)))))))))).------- ( -24.91, z-score = 0.13, R) >droAna3.scaffold_13047 1490783 91 + 1816235 -----------------UCCUGGCACCACCCGCCAUGACCCCCCCCCCCCCC------CUUUUUUUUCUGGUUCUGACUCUGGCGUGACAAAUACAUGAGGGGGGGGUCCAAUG -----------------...((((.......)))).((((((((((......------.........(..(........)..)((((.......)))).))))))))))..... ( -31.50, z-score = -1.01, R) >consensus GUUGGCCCC____C_GCUCCCAGGAUCCGCCACUUCCAUCCGGGGAAUAUCU_____CCCCCUUUUUCUGCUUCUGACUUUGGCGUGACAAAUGCAUGGGGCCAAUG_______ (((((((((................................((((.............))))....................((((.....))))..)))))))))........ (-15.84 = -18.50 + 2.67)

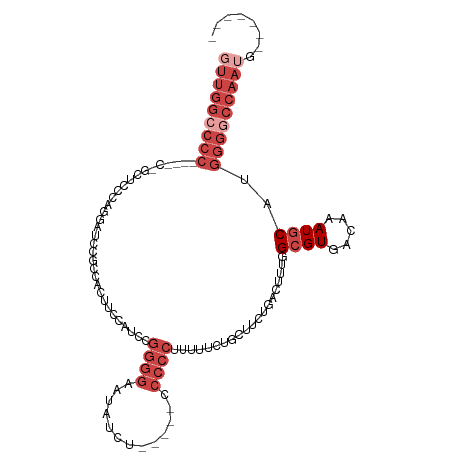
| Location | 13,982,193 – 13,982,291 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.46086 |
| G+C content | 0.56120 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -19.17 |
| Energy contribution | -22.12 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13982193 98 - 22422827 -------CAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCGU----GGGGCCAAC -------..((((((((((((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))))----)))))))). ( -46.50, z-score = -4.52, R) >droSim1.chrX_random 3745339 98 - 5698898 -------CAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCAG----GGGGCCAAC -------..((((((((.(((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))).----)))))))). ( -42.60, z-score = -3.40, R) >droSec1.super_20 601901 98 - 1148123 -------CAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG-----AGAUAUUCCCCGGAUGGAAGUGGCGGAUCCUGGGAGCAG----GGGGCCAAC -------..((((((((.(((.((((((((.(((..((.....)).......(((((-----(....))))))...)))..))))))))((....))))).----)))))))). ( -42.60, z-score = -3.40, R) >droYak2.chrX 8248527 107 - 21770863 -------CAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAGCAGCAGAAAAAGGGGGGAGAUAUAUAUUCCCCGGUUGAAAGUGGAGGUUCCUGGCAGCGGGUUUGGGGCCAAA -------..(((((((((.((..(((((...(((...(((((..(.......)((((((........)))))).)))))...)))((....)))))))...)).))))))))). ( -42.20, z-score = -2.25, R) >droEre2.scaffold_4690 5812938 97 + 18748788 -------CAUUGGUCCCAUGCAUUUGUCACGCCAAAGUCAGAAGCUGAAAAAGGGGG-----ACAUAUUCCCCGGAUGAAAGUGGUGGAGCCUGGGAGU-G----GGGGCCAAC -------..((((((((...(....((..(((((...(((....((.....))((((-----(.....)))))...)))...)))))..))....)...-.----)))))))). ( -34.90, z-score = -1.30, R) >droAna3.scaffold_13047 1490783 91 - 1816235 CAUUGGACCCCCCCCUCAUGUAUUUGUCACGCCAGAGUCAGAACCAGAAAAAAAAG------GGGGGGGGGGGGGGUCAUGGCGGGUGGUGCCAGGA----------------- .....((((((((((((.....((((.(.(....).).)))).((..........)------)...)))))))))))).(((((.....)))))...----------------- ( -37.20, z-score = -0.65, R) >consensus _______CAUUGGCCCCAUGCAUUUGUCACGCCAAAGUCAAAAGCAGAAAAAGGGGG_____AGAUAUUCCCCGGAUGAAAGUGGCGGAUCCUGGGAGC_G____GGGGCCAAC .........((((((((.(((.((((.(........).)))).))).....................(((((.((((...........)))).))))).......)))))))). (-19.17 = -22.12 + 2.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:42:00 2011