
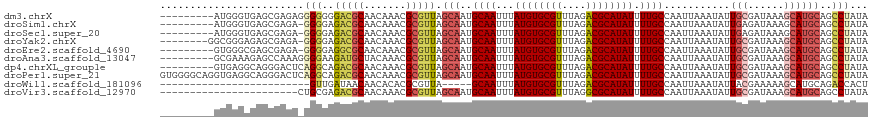
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,967,919 – 13,968,067 |
| Length | 148 |
| Max. P | 0.862458 |

| Location | 13,967,919 – 13,968,028 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.71 |
| Shannon entropy | 0.32609 |
| G+C content | 0.42436 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.81 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623786 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 13967919 109 - 22422827 ---------AUGGGUGAGCGAGAGGGGGGGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ---------((((((..(((....(.(....).)......)))....(((..((((...((((((((....)))))))).))))...........(((......)))))).)))))). ( -31.90, z-score = -1.52, R) >droSim1.chrX 10735774 108 - 17042790 ---------AUGGGUGAGCGAGA-GGGGAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGAGAUAAAGCAUGCAGCCUAUA ---------((((((..(((...-.....(((((.......))))).((...((((...((((((((....)))))))).))))((((.....)))).......)).))).)))))). ( -30.90, z-score = -2.17, R) >droSec1.super_20 587985 108 - 1148123 ---------AUGGGUGAGCGAGA-GGGGAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGAGAUAAAGCAUGCAGCCUAUA ---------((((((..(((...-.....(((((.......))))).((...((((...((((((((....)))))))).))))((((.....)))).......)).))).)))))). ( -30.90, z-score = -2.17, R) >droYak2.chrX 8233751 109 - 21770863 --------GGCGGGAGAGCGAGA-GGGGAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA --------(((............-.....(((((.......))))).(((..((((...((((((((....)))))))).))))...........(((......)))))).))).... ( -29.60, z-score = -1.09, R) >droEre2.scaffold_4690 5798536 108 + 18748788 ---------GUGGGCGAGCGAGA-GGGGAGGCGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ---------((((((........-.....(((((.......))))).(((..((((...((((((((....)))))))).))))...........(((......)))))).)))))). ( -32.60, z-score = -1.22, R) >droAna3.scaffold_13047 1476295 109 - 1816235 ---------GCGAAAGAGCCAAAGGGGAAGAUGCUACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ---------((......))....(((...(((((.......))))).(((..((((...((((((((....)))))))).))))...........(((......))))))..)))... ( -26.30, z-score = -0.26, R) >dp4.chrXL_group1e 6252854 109 + 12523060 ---------GUGAGGCAGGGACUCAGGCAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ---------..(((.......)))((((.(((((.......))))).(((..((((...((((((((....)))))))).))))...........(((......)))))).))))... ( -33.70, z-score = -2.21, R) >droPer1.super_21 387198 118 + 1645244 GUGGGGCAGGUGAGGCAGGGACUCAGGCAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ...((((...((((.......)))).((((((((.......))))).(((((((((...((((((((....)))))))).)))).........))))).........))).))))... ( -35.20, z-score = -1.21, R) >droWil1.scaffold_181096 8556553 88 - 12416693 -------------------------GUUGAUAACAACACACGCGUUA-----GCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUACGAAAAAGCAUGCAGACCACU -------------------------((((....))))....((((..-----((((...((((((((....)))))))).))))..............(......)))))........ ( -17.00, z-score = -1.00, R) >droVir3.scaffold_12970 10225272 95 + 11907090 -----------------------CUGCGAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGGCGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA -----------------------(((((.(((((.......))))).(((((((((..(((((((......)))))))..)))).........))))).........)))))...... ( -28.70, z-score = -1.55, R) >consensus _________GUGGGUGAGCGAGA_GGGGAGACGCAACAAACGCGUUAGCAAUGCAAUUUAUGUGCGUUUAGACGCAUAUUUUGCCAAUUAAAUAUUGCGAUAAAGCAUGCAGCCUAUA ........................(((..(((((.......))))).(((..((((...((((((((....)))))))).))))...........(((......))))))..)))... (-21.23 = -21.81 + 0.58)


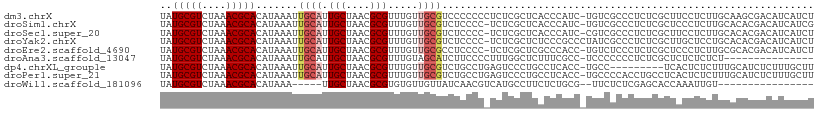
| Location | 13,967,959 – 13,968,067 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Shannon entropy | 0.63656 |
| G+C content | 0.51263 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -7.68 |
| Energy contribution | -8.05 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
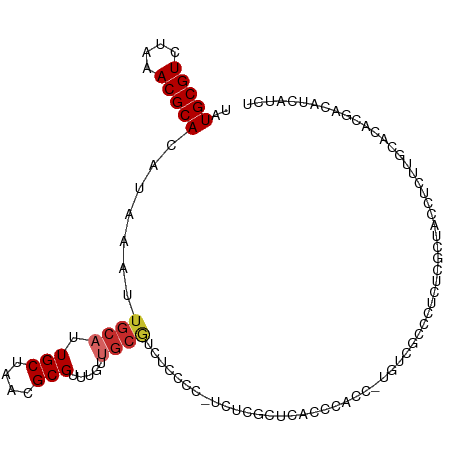
>dm3.chrX 13967959 108 + 22422827 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCCCCCCCUCUCGCUCACCCAUC-UGUCGCCCUCUCGCUUCCUCUUGCAAGCGACAUCAUCU ...(((.(((((((((.......)))...((....)))))))..(((...........))).........-.).)))....((((((.(....).))))))........ ( -19.70, z-score = -1.93, R) >droSim1.chrX 10735814 107 + 17042790 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUCCCC-UCUCGCUCACCCAUC-UGUCGCCCUCUCGCUCCCUCUUGCACACGACAUCAUCG .(((((..((((((((.......)))...((....))))))).)))))......-...............-(((((.......((........))...)))))...... ( -15.90, z-score = -0.87, R) >droSec1.super_20 588025 107 + 1148123 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUCCCC-UCUCGCUCACCCAUC-CGUCGCCCUCUCGCUUCCUCUUGCACACGACAUCAUCU .(((((..((((((((.......)))...((....))))))).)))))......-...............-.((((.......((........))...))))....... ( -15.60, z-score = -0.82, R) >droYak2.chrX 8233791 108 + 21770863 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUCCCC-UCUCGCUCUCCCGCCCUAUCGCCCUCUCGCUUGCUCCUGCACACGACAUCAUCU ..(((((....)))))........(((.(((....))).....)))(((.....-....((......((......))......)).(((....)))...)))....... ( -17.50, z-score = -1.05, R) >droEre2.scaffold_4690 5798576 107 - 18748788 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGCCUCCCC-UCUCGCUCGCCCACC-UGUCUCCCUCUCGCUCCCUCUUGCGCACGACAUCAUCU ...((((.((((((((.......)))...((....)))))))..))))......-...............-((((.......(((........)))...))))...... ( -17.10, z-score = -1.09, R) >droAna3.scaffold_13047 1476335 93 + 1816235 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUAGCAUCUUCCCCUUUGGCUCUUUCGCC-UCCCCCCCUCUCGCUCUCUCUCU--------------- ..(((((....))))).......((((.(((....)))..))))((............(((......)))-............)).........--------------- ( -15.75, z-score = -2.64, R) >dp4.chrXL_group1e 6252894 99 - 12523060 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUGCCUGAGUCCCUGCCUCACC-UGCC---------UCACUCUCUUUGCAUCUCUUUGCUU ..(((((....))))).......((((..((..(((((.....)))))..))..((((....((......-.)).---------..))))....))))........... ( -19.80, z-score = -1.65, R) >droPer1.super_21 387238 108 - 1645244 UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUGCCUGAGUCCCUGCCUCACC-UGCCCCACCUGCCUCACUCUCUUUGCAUCUCUUUGCUU ..(((((....)))))........(((..((..(((((.....)))))..)).((((.......))))..-))).....................(((......))).. ( -19.50, z-score = -1.27, R) >droWil1.scaffold_181096 8556593 86 + 12416693 UAUGCGUCUAAACGCACAUAAA-----UUGCUAACGCGUGUGUUGUUAUCAACGUCAUGCCUUCUCUGCG--UUCUCUCGAGCACCAAAUUGU---------------- ..(((((....)))))......-----..((....((((((((((....))))).))))).......))(--(((....))))..........---------------- ( -19.50, z-score = -1.46, R) >consensus UAUGCGUCUAAACGCACAUAAAUUGCAUUGCUAACGCGUUUGUUGCGUCUCCCC_UCUCGCUCACCCACC_UGUCGCCCUCUCGCUACCUCUUGCACACGACAUCAUCU .(((((..(((((((....................))))))).)))))............................................................. ( -7.68 = -8.05 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:57 2011