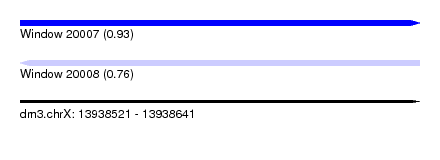
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,938,521 – 13,938,641 |
| Length | 120 |
| Max. P | 0.931875 |

| Location | 13,938,521 – 13,938,641 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.21 |
| Shannon entropy | 0.08091 |
| G+C content | 0.42920 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.931875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
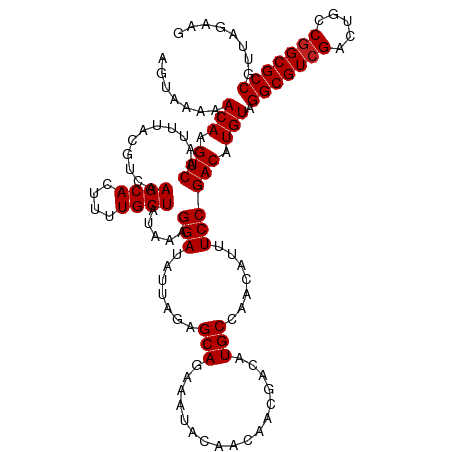
>dm3.chrX 13938521 120 + 22422827 CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.68, R) >droSim1.chrX 10722351 120 + 17042790 CUUCUAACGGCGCGGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....(((((((((.((((.((((((......))))))...)))).))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -31.20, z-score = -1.24, R) >droSec1.super_10504 687 120 + 1366 CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCCUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.10, z-score = -2.40, R) >droYak2.chrX 8219601 120 + 21770863 CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.68, R) >droEre2.scaffold_4690 5784557 120 - 18748788 CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.68, R) >droAna3.scaffold_13047 1459666 120 + 1816235 CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.68, R) >dp4.chrXL_group1e 6233349 120 - 12523060 CCUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.66, R) >droPer1.super_21 367974 120 - 1645244 CCUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ....((((((((((((((.((((((......))))))...)))))))..)))))))..((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -34.90, z-score = -2.66, R) >droWil1.scaffold_181096 3878763 120 - 12416693 CUUGCAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ...(((((((((((((((.((((((......))))))...)))))))..)))))))).((((((.((((.(((......))).)))).)))))).....(((((..........))))). ( -38.10, z-score = -3.06, R) >droVir3.scaffold_12970 1704902 113 + 11907090 -------CGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCCUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU -------....(((((((.((((((......))))))...)))))))(((((......((((((.((((.(((......))).)))).))))))..)))))................... ( -30.70, z-score = -1.74, R) >droMoj3.scaffold_6359 2906157 113 + 4525533 -------CGGCGCCGCUAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCCUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU -------.((((.((.....)).)))).(((.((((((.(((....))).)))..(((((((((.((((.(((......))).)))).))))))..))).......))).)))....... ( -27.90, z-score = -1.36, R) >droGri2.scaffold_15081 1452142 113 + 4274704 -------CGGCGCCGCCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCCUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU -------.(((...)))((((((.(((.((((.......)))).)))(((((......((((((.((((.(((......))).)))).))))))..)))))...)))))).......... ( -29.30, z-score = -1.71, R) >consensus CUUCUAACGGCGCCGGCAGUCGACGCCUACAUGUCGGAAAUGUUGGCAUGUCGUUGUUGUAUUUCUGCUCUAAUAUCCUUUAUAGCAAAAGUGCUUGACGUAAAUUGACUUGUUUUUACU ...........(((((((.((((((......))))))...)))))))(((((......((((((.((((.(((......))).)))).))))))..)))))................... (-29.48 = -29.65 + 0.17)
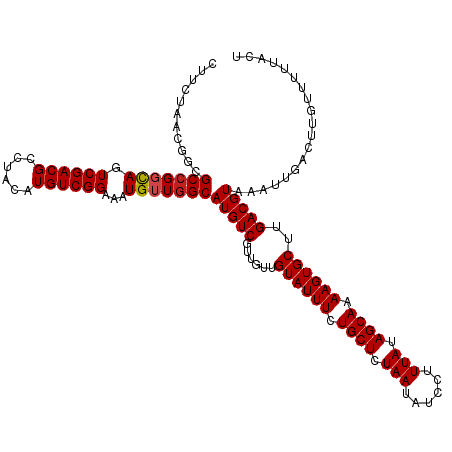
| Location | 13,938,521 – 13,938,641 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.21 |
| Shannon entropy | 0.08091 |
| G+C content | 0.42920 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -25.51 |
| Energy contribution | -25.59 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.755763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13938521 120 - 22422827 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -1.09, R) >droSim1.chrX 10722351 120 - 17042790 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCCGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).((((.((......))))))........ ( -21.77, z-score = -0.02, R) >droSec1.super_10504 687 120 - 1366 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAGGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((................(((((((.(((......))).)))))))..........((.(((....))).)).))).))).(((((((.....)))))))........ ( -26.50, z-score = -0.96, R) >droYak2.chrX 8219601 120 - 21770863 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -1.09, R) >droEre2.scaffold_4690 5784557 120 + 18748788 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -1.09, R) >droAna3.scaffold_13047 1459666 120 - 1816235 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -1.09, R) >dp4.chrXL_group1e 6233349 120 + 12523060 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAGG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -0.79, R) >droPer1.super_21 367974 120 + 1645244 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAGG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ ( -25.97, z-score = -0.79, R) >droWil1.scaffold_181096 3878763 120 + 12416693 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUGCAAG .((((..(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....))))))).))))... ( -28.17, z-score = -1.33, R) >droVir3.scaffold_12970 1704902 113 - 11907090 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAAGGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCG------- .......(((.(((................(((((((.(((......))).))))))).........(((.(((....))).)))))).))).(((((((.....))))))).------- ( -29.80, z-score = -2.64, R) >droMoj3.scaffold_6359 2906157 113 - 4525533 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAAGGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUAGCGGCGCCG------- .......(((.(((................(((((((.(((......))).))))))).........(((.(((....))).)))))).))).(((((((.....))))))).------- ( -29.60, z-score = -2.95, R) >droGri2.scaffold_15081 1452142 113 - 4274704 AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAAGGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGGCGGCGCCG------- .......(((.(((................(((((((.(((......))).))))))).........(((.(((....))).)))))).))).(((((((.....))))))).------- ( -29.60, z-score = -2.42, R) >consensus AGUAAAAACAAGUCAAUUUACGUCAAGCACUUUUGCUAUAAAGGAUAUUAGAGCAGAAAUACAACAACGACAUGCCAACAUUUCCGACAUGUAGGCGUCGACUGCCGGCGCCGUUAGAAG .......(((.(((...........((((....)))).....(((.......(((.................))).......)))))).))).(((((((.....)))))))........ (-25.51 = -25.59 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:54 2011