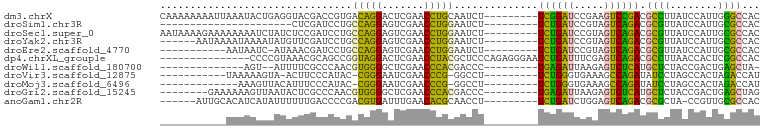
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,911,250 – 13,911,341 |
| Length | 91 |
| Max. P | 0.997163 |

| Location | 13,911,250 – 13,911,341 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 55.29 |
| Shannon entropy | 0.86397 |
| G+C content | 0.50251 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -15.05 |
| Energy contribution | -11.56 |
| Covariance contribution | -3.49 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
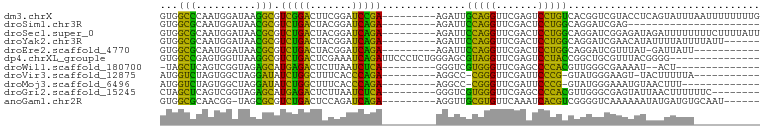
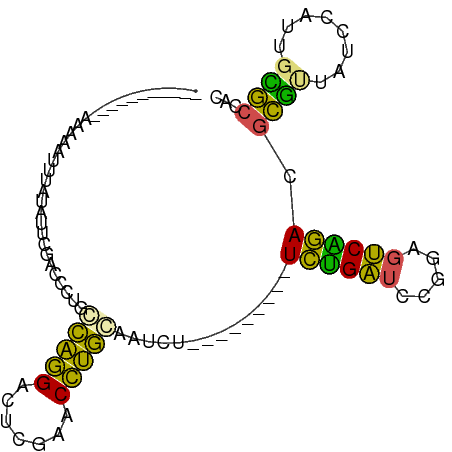
>dm3.chrX 13911250 91 + 22422827 GUGGCCCAAUGGAUAAGGCGUCGGACUUCGGAUCCGA---------AGAUUGCAGGUUCGAGUCCUGUCACGGUCGUACCUCAGUAUUUAAUUUUUUUUG ..((((.....((((.(((.((((((((..((((...---------.))))..))))))))))).))))..))))((((....))))............. ( -22.60, z-score = 0.09, R) >droSim1.chr3R 17292399 69 + 27517382 GUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGAG---------------------- ...(((..........))).((((((....).)))))---------.(((((((((.......)))))...))))...---------------------- ( -18.10, z-score = 0.23, R) >droSec1.super_0 17862462 91 + 21120651 GUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGGAGAUAGAUUUUUUUUCUUUUAUU ...(((..........))).((((((....).)))))---------.(((((((((.......)))))...))))(((((........)))))....... ( -19.80, z-score = 0.70, R) >droYak2.chr3R 20795027 85 - 28832112 GUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGAACAUAUUUUAUUUUAUU------ .........((((...(...((((((....).)))))---------.)..)))).((((((.(((.....)))))))))...............------ ( -22.10, z-score = -1.28, R) >droEre2.scaffold_4770 17615425 79 + 17746568 GUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGUUUAU-GAUUAUU----------- ..(((((..........)))))........(((((..---------.(((((((((.......)))))...)))).....)-))))...----------- ( -19.20, z-score = 0.13, R) >dp4.chrXL_group1e 6235814 85 + 12523060 GUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAGUCCUACCGGCUGCGUUUACGGGG--------------- (..((((.((((...((....))((((((((....(.(((((...)))))).....))))))))))))))))..)..........--------------- ( -31.30, z-score = -0.81, R) >droWil1.scaffold_180700 738053 74 - 6630534 -UAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCA---------GGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAU--ACU-------------- -..((((((.(....((.(.(((((.......)))))---------.).))(((((.......))))))))))))......--...-------------- ( -23.10, z-score = -1.03, R) >droVir3.scaffold_12875 4787947 77 + 20611582 AUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGA---------AGGCC-CGGGUUCGAUUCCCG-GUAUGGGAAGU-UACUUUUUA----------- .......(((((((.((((.((.(((((((.....))---------)))))-.))))))...((((.-....)))))))-)))).....----------- ( -23.60, z-score = -0.31, R) >droMoj3.scaffold_6496 1583829 76 - 26866924 AUGGUCUAGUGGCUAGGAUAUCUGGCUUUCACCCAGA---------AGGCC-CGGGUUCGAUUCCCG-GUAUGGGAAAUGUAACUUU------------- .(((((....)))))........(((((((.....))---------)))))-.(((((((.(((((.-....))))).)).))))).------------- ( -22.80, z-score = -0.14, R) >droGri2.scaffold_15245 207054 83 + 18325388 CUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCA---------GGGUCGUGGGUUCGAGCCCCACGUUGGGCGAGUAUUAACUUUUUUC-------- ...((((........((((....((((((.......)---------)))))....))))..((((......)))))))).............-------- ( -24.00, z-score = -0.81, R) >anoGam1.chr2R 5380054 84 - 62725911 GUGGCGCAACGG-UAGCGCGUCUGACUCCAGAUCAGA---------AGGUUGCGUGUUCAAAUCACGUCGGGGUCAAAAAAUAUGAUGUGCAAU------ ............-..((((((((((((((.((((...---------.))))(((((.......))))).)))))))........)))))))...------ ( -25.40, z-score = -1.38, R) >consensus GUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGA_________AGAUUGCAGGUUCGACUCCUGGCAGGGUCGAAUAUAAAUUUUU___________ ...(((..........))).(((((.......)))))..............(((((.......)))))................................ (-15.05 = -11.56 + -3.49)
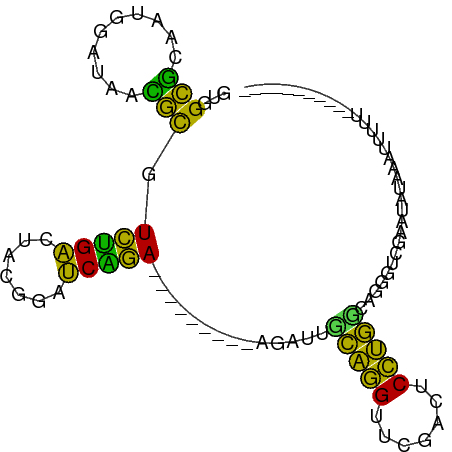
| Location | 13,911,250 – 13,911,341 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 55.29 |
| Shannon entropy | 0.86397 |
| G+C content | 0.50251 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -16.81 |
| Energy contribution | -14.35 |
| Covariance contribution | -2.46 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13911250 91 - 22422827 CAAAAAAAAUUAAAUACUGAGGUACGACCGUGACAGGACUCGAACCUGCAAUCU---------UCGGAUCCGAAGUCCGACGCCUUAUCCAUUGGGCCAC .................((((((.(((((......))..))).)))).))....---------((((((.....)))))).((((........))))... ( -21.00, z-score = -0.92, R) >droSim1.chr3R 17292399 69 - 27517382 ----------------------CUCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCAC ----------------------...(((....(((((.......))))).))).---------((((((.....)))))).((((........))))... ( -17.90, z-score = -0.65, R) >droSec1.super_0 17862462 91 - 21120651 AAUAAAAGAAAAAAAAUCUAUCUCCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCAC .........................(((....(((((.......))))).))).---------((((((.....)))))).((((........))))... ( -17.90, z-score = -0.88, R) >droYak2.chr3R 20795027 85 + 28832112 ------AAUAAAAUAAAAUAUGUUCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCAC ------...............(((((((((.....))).)))))).(((.....---------((((((.....))))))((((........))))))). ( -21.20, z-score = -2.08, R) >droEre2.scaffold_4770 17615425 79 - 17746568 -----------AAUAAUC-AUAAACGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCAC -----------.......-......(((....(((((.......))))).))).---------((((((.....)))))).((((........))))... ( -17.90, z-score = -1.01, R) >dp4.chrXL_group1e 6235814 85 - 12523060 ---------------CCCCGUAAACGCAGCCGGUAGGACUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCAC ---------------.............(((((((((.......)))))........((((..((((((.....))))))..)))).......))))... ( -24.20, z-score = -0.93, R) >droWil1.scaffold_180700 738053 74 + 6630534 --------------AGU--AUUUUCGCCCAACGUGGGGCUCGAACCCACGACCC---------UGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUA- --------------...--............((((((.......))))))....---------((((((.....)))))).((((........))))..- ( -22.50, z-score = -2.17, R) >droVir3.scaffold_12875 4787947 77 - 20611582 -----------UAAAAAGUA-ACUUCCCAUAC-CGGGAAUCGAACCCG-GGCCU---------UCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAU -----------.........-..........(-((((.......))))-)....---------(((((.(....))))))....((((...))))..... ( -16.60, z-score = 0.06, R) >droMoj3.scaffold_6496 1583829 76 + 26866924 -------------AAAGUUACAUUUCCCAUAC-CGGGAAUCGAACCCG-GGCCU---------UCUGGGUGAAAGCCAGAUAUCCUAGCCACUAGACCAU -------------...(((....(((((....-.)))))...)))..(-(....---------(((((.(....))))))....((((...)))).)).. ( -17.40, z-score = -0.08, R) >droGri2.scaffold_15245 207054 83 - 18325388 --------GAAAAAAGUUAAUACUCGCCCAACGUGGGGCUCGAACCCACGACCC---------UGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAG --------.......................((((((.......))))))....---------((((((.....)))))).((((........))))... ( -22.50, z-score = -2.32, R) >anoGam1.chr2R 5380054 84 + 62725911 ------AUUGCACAUCAUAUUUUUUGACCCCGACGUGAUUUGAACACGCAACCU---------UCUGAUCUGGAGUCAGACGCGCUA-CCGUUGCGCCAC ------.((((...(((.......))).......(((.......)))))))...---------((((((.....)))))).((((..-.....))))... ( -17.20, z-score = -0.70, R) >consensus ___________AAAAAUUUAUAUUCGACCCUGCCAGGACUCGAACCUGCAAUCU_________UCUGAUCCGGAGUCAGACGCGUUAUCCAUUGCGCCAC ................................(((((.......)))))..............((((((.....)))))).((((........))))... (-16.81 = -14.35 + -2.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:51 2011