| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,371,221 – 11,371,331 |
| Length | 110 |
| Max. P | 0.996583 |

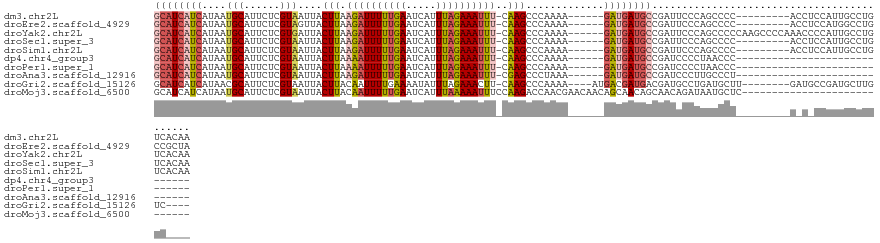
| Location | 11,371,221 – 11,371,331 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.43744 |
| G+C content | 0.37784 |
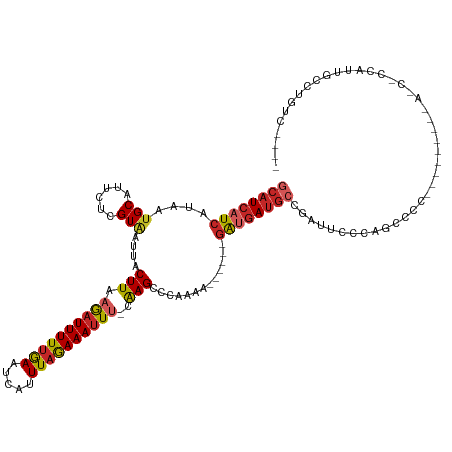
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -12.18 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11371221 110 - 23011544 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUUCCCAGCCCC---------ACCUCCAUUGCCUGUCACAA ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))..............---------.................... ( -18.80, z-score = -2.32, R) >droEre2.scaffold_4929 12568546 110 + 26641161 GCAUCAUCAUAAUGCAUUCUCGUAGUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUUCCCAGCCCC---------ACCUCCAUGGCCUGCCGCUA ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))........(((..(---------(((.....))..))..))). ( -22.00, z-score = -2.00, R) >droYak2.chr2L 7767527 119 - 22324452 GCAUCAUCAUAAUGCAUUCUCGUGAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUUCCCAGCCCCCAAGCCCCAAACCCCAUUGCCUGUCACAA ((((((((.((((.(((....)))))))(((.((((((((((.....))))))))))-.))).......------))))))))........................................... ( -18.80, z-score = -1.83, R) >droSec1.super_3 6765750 110 - 7220098 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUUCCCAGCCCC---------ACCUCCAUUGCCUGUCACAA ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))..............---------.................... ( -18.80, z-score = -2.32, R) >droSim1.chr2L 11177320 110 - 22036055 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUUCCCAGCCCC---------ACCUCCAUUGCCUGUCACAA ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))..............---------.................... ( -18.80, z-score = -2.32, R) >dp4.chr4_group3 6814898 90 - 11692001 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAAAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUCCCCUAACCC----------------------------- ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))..............----------------------------- ( -18.70, z-score = -4.67, R) >droPer1.super_1 8262663 90 - 10282868 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAAAUUUUUGAAUCAUUUAGAAAUUU-CAAGCCCAAAA------GAUGAUGCCGAUCCCCUAACCC----------------------------- ((((((((....(((......)))....(((.((((((((((.....))))))))))-.))).......------))))))))..............----------------------------- ( -18.70, z-score = -4.67, R) >droAna3.scaffold_12916 12082071 90 + 16180835 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU-CGAGCCCUAAA------GAUGAUGCCGAUCCCUUGCCCU----------------------------- ((((((((....(((......)))....((..((((((((((.....))))))))))-..)).......------))))))))..............----------------------------- ( -20.10, z-score = -3.37, R) >droGri2.scaffold_15126 7189596 109 + 8399593 GCAUCAUCAUAACGCAUUCUCGUAAUUACUUACAAUUUUGAAAAUAUUUAGAAACUU-CAAGCCCAAAA----AUGACGAUGACGAUGCCUGAUGCUU--------GAUGCCGAUGCUUGUC---- ((((((.(((...(((((.((((..(((........((((((.............))-)))).......----.)))..)))).)))))...)))..)--------)))))...........---- ( -19.81, z-score = -1.05, R) >droMoj3.scaffold_6500 5022945 98 + 32352404 GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUACAAUUUUUGAAUCAUUUAAAAAUUUCCAAGACCAACGAACAACAGCAACAGCAACAGAUAAUGCUC---------------------------- ((((.(((...........((((.....(((..(((((((((.....)))))))))...)))....))))......((....))....))).))))..---------------------------- ( -13.80, z-score = -2.39, R) >consensus GCAUCAUCAUAAUGCAUUCUCGUAAUUACUUAAGAUUUUUGAAUCAUUUAGAAAUUU_CAAGCCCAAAA______GAUGAUGCCGAUUCCCAGCCCC_________A_C_CCAUUGCCUGUC____ ((((((((....(((......)))....(((..(((((((((.....)))))))))...))).............))))))))........................................... (-12.18 = -12.60 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:13 2011