
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,910,222 – 13,910,329 |
| Length | 107 |
| Max. P | 0.998035 |

| Location | 13,910,222 – 13,910,329 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.61 |
| Shannon entropy | 0.64313 |
| G+C content | 0.51882 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -28.77 |
| Energy contribution | -24.63 |
| Covariance contribution | -4.14 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
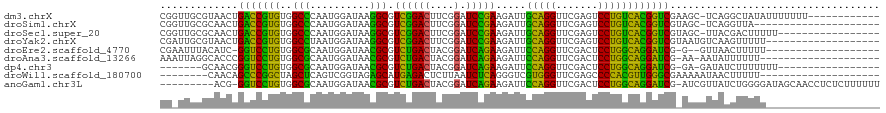
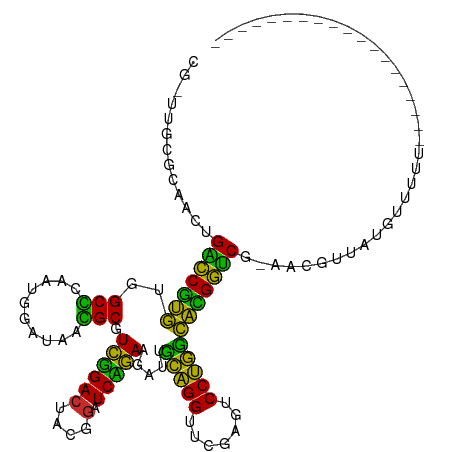
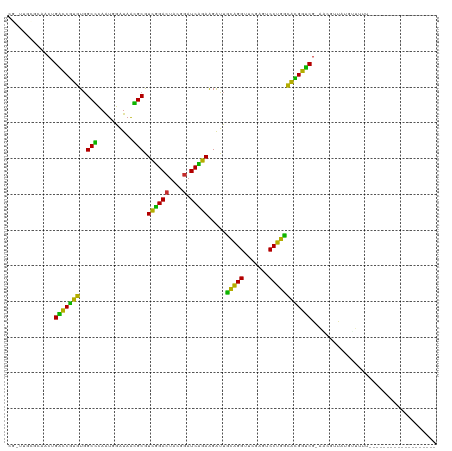
>dm3.chrX 13910222 107 + 22422827 CGGUUGCGUAACUGACCGUGUGGCCCAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGAAGC-UCAGGCUAUAUUUUUUU------------ .(.(((.((...((((((((.(((((.........)).)))(((((.((((((.(.......).)))))))))))...))))))))..))-.))).)...........------------ ( -32.90, z-score = -0.19, R) >droSim1.chrX 6290597 98 - 17042790 CGGUUGCGCAACUGACCGUGUGGCCCAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGUAGC-UCAGGUUA--------------------- ...(((.((.((.(((((((.(((((.........)).)))(((((.((((((.(.......).)))))))))))...))))))))).))-.)))....--------------------- ( -33.80, z-score = -0.43, R) >droSec1.super_20 543807 102 + 1148123 CGGUUGCGCAACUGACCGUGUGGCCCAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGUAGC-UUACGACUUUUU----------------- .(((((.((.((.(((((((.(((((.........)).)))(((((.((((((.(.......).)))))))))))...))))))))).))-...)))))....----------------- ( -34.80, z-score = -0.76, R) >droYak2.chrX 8196753 101 + 21770863 CGAUUGCGUAACUGACCGUGUGGCCUAAUGGAUAAGGCGUCGGACUUCGGAUCCGAAGAUUGCAGGUUCGAGUCCUGUCACGGUCGUAAUGUCAAGUUUUU------------------- ...((((((.((.(((((((..((((........)))).((((((....).))))).....(((((.......)))))))))))))).))).)))......------------------- ( -34.80, z-score = -1.91, R) >droEre2.scaffold_4770 17614591 97 + 17746568 CGAAUUUACAUC-GGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-G--GUUAACUUUUU------------------- ..........((-(((((((((((((..........))))).......(((((....)))))((((.......)))))))))))))-)--...........------------------- ( -29.40, z-score = -1.25, R) >droAna3.scaffold_13266 1140290 98 - 19884421 AAAUUAGGCACCCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-AA-AAUAUUUUUU-------------------- ............((((((((((((((..........))))).......(((((....)))))((((.......)))))))))))))-..-..........-------------------- ( -28.70, z-score = -1.10, R) >dp4.chr3 1788224 94 - 19779522 -------GCAACGGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-GA-GAUAUCUUUUUUU----------------- -------....(.(((((((((((((..........))))).......(((((....)))))((((.......)))))))))))).-).-.............----------------- ( -27.90, z-score = -0.52, R) >droWil1.scaffold_180700 724594 92 - 6630534 --------CAACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAAAAUAACUUUUU-------------------- --------.....(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))...............-------------------- ( -32.90, z-score = -2.41, R) >anoGam1.chr3L 7656875 109 - 41284009 ---------ACG-GGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCG-AUCGUUAUCUGGGGAUAGCAACCUCUCUUUUUU ---------...-(((((((((((((..........))))).......(((((....)))))((((.......)))))))))))).-...((((((....)))))).............. ( -32.70, z-score = -0.17, R) >consensus CG_UUGCGCAACUGACCGUGUGGCCCAAUGGAUAACGCGUCGGACUACGGAUCAGAAGAUUGCAGGUUCGAGUCCUGGCACGGUCG_AACGUUAUGUUUUU___________________ .............(((((((..(((..........))).(((((.......))))).....(((((.......))))))))))))................................... (-28.77 = -24.63 + -4.14)
| Location | 13,910,222 – 13,910,329 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.61 |
| Shannon entropy | 0.64313 |
| G+C content | 0.51882 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -24.11 |
| Energy contribution | -21.28 |
| Covariance contribution | -2.83 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
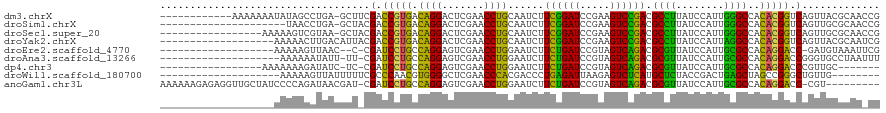
>dm3.chrX 13910222 107 - 22422827 ------------AAAAAAAUAUAGCCUGA-GCUUCGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUGGGCCACACGGUCAGUUACGCAACCG ------------...........((...(-(((..(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))))))..))..... ( -31.90, z-score = -2.28, R) >droSim1.chrX 6290597 98 + 17042790 ---------------------UAACCUGA-GCUACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUGGGCCACACGGUCAGUUGCGCAACCG ---------------------.....((.-((.(((((((((.((((.......))))......((((((.....)))))).((((........))))..))))))).)).)).)).... ( -32.30, z-score = -2.26, R) >droSec1.super_20 543807 102 - 1148123 -----------------AAAAAGUCGUAA-GCUACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUGGGCCACACGGUCAGUUGCGCAACCG -----------------.....(.(((((-.((..(((((((.((((.......))))......((((((.....)))))).((((........))))..)))))))))))))))..... ( -32.10, z-score = -1.73, R) >droYak2.chrX 8196753 101 - 21770863 -------------------AAAAACUUGACAUUACGACCGUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCCGACGCCUUAUCCAUUAGGCCACACGGUCAGUUACGCAAUCG -------------------......(((.....(((((((((.((((.......))))......((((((.....)))))).((((........))))..))))))).))....)))... ( -28.80, z-score = -2.59, R) >droEre2.scaffold_4770 17614591 97 - 17746568 -------------------AAAAAGUUAAC--C-CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC-GAUGUAAAUUCG -------------------...........--.-((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)-)........... ( -25.60, z-score = -1.81, R) >droAna3.scaffold_13266 1140290 98 + 19884421 --------------------AAAAAAUAUU-UU-CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGGGUGCCUAAUUU --------------------..........-((-((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)))).......... ( -26.50, z-score = -1.08, R) >dp4.chr3 1788224 94 + 19779522 -----------------AAAAAAAGAUAUC-UC-CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCCGUUGC------- -----------------.............-..-((.((((((((((.......))))).....((((((.....)))))).((((........))))..)))))..))....------- ( -23.70, z-score = -0.95, R) >droWil1.scaffold_180700 724594 92 + 6630534 --------------------AAAAAGUUAUUUUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUUG-------- --------------------...............((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).....-------- ( -31.00, z-score = -2.65, R) >anoGam1.chr3L 7656875 109 + 41284009 AAAAAAGAGAGGUUGCUAUCCCCAGAUAACGAU-CGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACC-CGU--------- ..........(((((.((((....)))).))))-)..((((((((((.......))))).....((((((.....)))))).((((........))))..)))))..-...--------- ( -29.80, z-score = -1.18, R) >consensus ___________________AAAAACAUAACGUU_CGACCCUGACAGGACUCGAACCUGCAAUCUUCGGAUCCGAAGUCAGACGCCUUAUCCAUUGCGCCACACGGCCAGUUGCGCAA_CG .....................................(((((.((((.......))))......((((((.....)))))).((((........))))..)))))............... (-24.11 = -21.28 + -2.83)
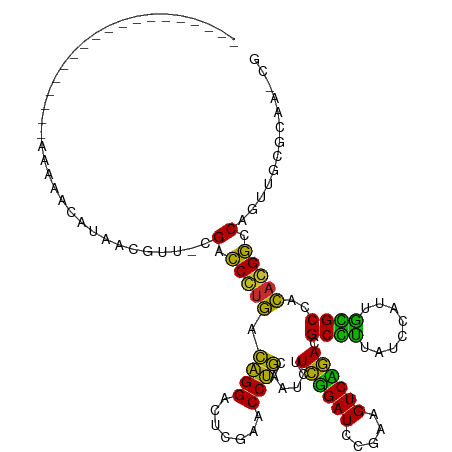
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:49 2011