| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,908,833 – 13,908,924 |
| Length | 91 |
| Max. P | 0.986851 |

| Location | 13,908,833 – 13,908,924 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.03 |
| Shannon entropy | 0.61000 |
| G+C content | 0.51453 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -17.11 |
| Energy contribution | -16.11 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
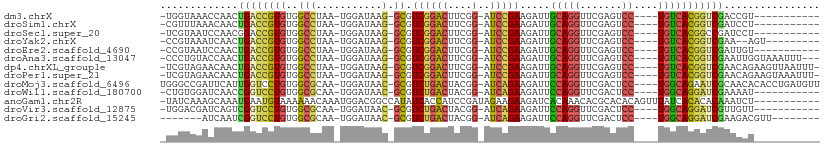
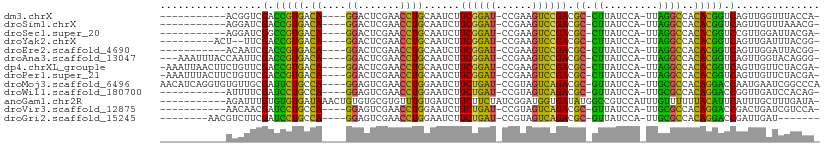
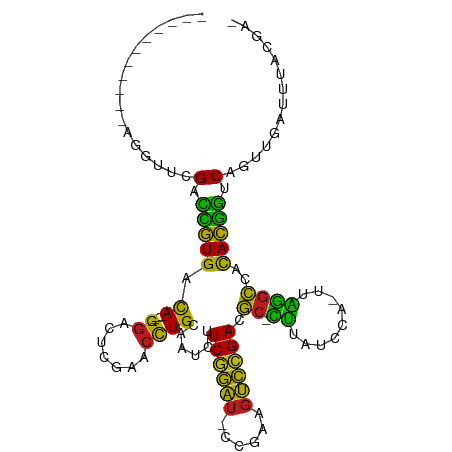
>dm3.chrX 13908833 91 + 22422827 -UGGUAAACCAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGACCGU----------- -(((....)))..((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----))))))))))).....----------- ( -33.50, z-score = -1.56, R) >droSim1.chrX 6289184 91 - 17042790 -CGUUUAAACAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAUCCU----------- -............((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----))))))))))).....----------- ( -31.50, z-score = -1.54, R) >droSec1.super_20 542450 91 + 1148123 -UCGUAAUCCAACGGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGCCGAUCCU----------- -...........(((.(((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----))))))))))).....----------- ( -32.00, z-score = -0.84, R) >droYak2.chrX 8195333 91 + 21770863 -CCGUAAAUCAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAA--AGU--------- -............((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----)))))))))))..--...--------- ( -31.50, z-score = -1.77, R) >droEre2.scaffold_4690 5760061 91 - 18748788 -CCGUAAUCCAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAUUGU----------- -........(((.((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----))))))))))).))).----------- ( -33.30, z-score = -1.85, R) >droAna3.scaffold_13047 1435200 99 + 1816235 -CCCUGUACCAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAAUUGGUAAAUUU--- -.....((((((.((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----)))))))))))..)))))).....--- ( -37.60, z-score = -2.81, R) >dp4.chrXL_group1e 6208347 101 - 12523060 -UCGUAGAACAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAACAGAAGUUAAUUU- -............((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----)))))))))))...............- ( -31.50, z-score = -1.14, R) >droPer1.super_21 341693 101 - 1645244 -UCGUAGAACAACUGACCGUGUGGCCUAA-UGGAUAAG-GCGUCGGACUUCGG-AUCCGAAGAUUGCAGGUUCGAGUCC----UGUCACGGUCGAACAGAAGUAAAUUU- -............((((((((..((((..-......))-)).((((((....)-.))))).....(((((.......))----)))))))))))...............- ( -31.50, z-score = -1.24, R) >droMoj3.scaffold_6496 1573678 103 - 26866924 UGGGCCGAUUCAUUGGUCCUGUGGCGCAA-UGGAUAAC-GCGUCUGACUACGG-AUCAGAAGAUUCCAGGUUCGACUCC----UGGCAGAAUCGCAACACACCUGAUGUU .(((((((....)))))))((((..((..-((((...(-...((((((....)-.))))).)..))))(((((..(...----..)..)))))))..))))......... ( -32.10, z-score = -0.65, R) >droWil1.scaffold_180700 720069 91 - 6630534 -CUGUGGAUCAACCGGUCCUGUGGCGCAA-UGGAUAAC-GCGUCUGACUACGG-AUCAGAAGAUUCCAGGUUCGACUCC----UGGCAGGAUCGAAAAU----------- -............((((((((((((((..-........-))))).......((-(((....)))))((((.......))----))))))))))).....----------- ( -28.70, z-score = -0.89, R) >anoGam1.chr2R 11360139 98 + 62725911 -UAUCAAAGCAAAUGAAUGUAAAAAACAAAUGGACGGCCAUAUCACCAUCCGAUAGAAGAAGAUCACAAACACGCACACAGUUUAUCACACACAAAUCU----------- -............((..(((.(.((((....(((.((........)).)))(((........)))...............)))).).)))..)).....----------- ( -8.60, z-score = 0.22, R) >droVir3.scaffold_12875 4777428 91 + 20611582 -UGGACGAUCAGUCGGUCCUGUGGCGCAA-UGGAUAAC-GCGUCUGACUACGG-AUCAGAAGAUUCCAGGUUCGACUCC----UGGCAGGAUCGUUGUU----------- -..((((((((((((..((((.(((((..-........-))))).......((-(((....)))))))))..)))))..----......)))))))...----------- ( -29.90, z-score = -0.46, R) >droGri2.scaffold_15245 18116430 88 - 18325388 -------AUCAAUCGGUCCUGUGGCGCAA-UGGAUAAC-GCGUCUGACUACGG-AUCAGAAGAUUCCAGGUUCGACUCC----UGGCAGGAUCGAAGACGUU-------- -------.....(((((((((((((((..-........-))))).......((-(((....)))))((((.......))----)))))))))))).......-------- ( -30.30, z-score = -1.51, R) >consensus _UCGUAAAUCAACUGACCGUGUGGCCUAA_UGGAUAAG_GCGUCGGACUUCGG_AUCCGAAGAUUGCAGGUUCGAGUCC____UGUCACGGUCGAACAU___________ .............((((((((.........(((((.......(((((........))))).........)))))............))))))))................ (-17.11 = -16.11 + -1.00)
| Location | 13,908,833 – 13,908,924 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.03 |
| Shannon entropy | 0.61000 |
| G+C content | 0.51453 |
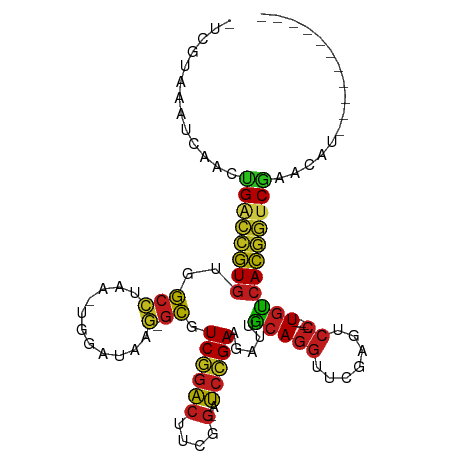
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -15.42 |
| Energy contribution | -12.82 |
| Covariance contribution | -2.59 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13908833 91 - 22422827 -----------ACGGUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGGUUUACCA- -----------.(((..(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..)))))))..)))........- ( -30.30, z-score = -1.66, R) >droSim1.chrX 6289184 91 + 17042790 -----------AGGAUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGUUUAAACG- -----------......(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))).(((.....))).- ( -29.40, z-score = -2.12, R) >droSec1.super_20 542450 91 - 1148123 -----------AGGAUCGGCCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCCGUUGGAUUACGA- -----------......(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..)))))))(((......))).- ( -28.50, z-score = -0.41, R) >droYak2.chrX 8195333 91 - 21770863 ---------ACU--UUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGAUUUACGG- ---------...--...(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))).............- ( -28.60, z-score = -2.27, R) >droEre2.scaffold_4690 5760061 91 + 18748788 -----------ACAAUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGGAUUACGG- -----------.((((.(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))).))))........- ( -31.70, z-score = -2.78, R) >droAna3.scaffold_13047 1435200 99 - 1816235 ---AAAUUUACCAAUUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGGUACAGGG- ---.....((((((((.(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))))))))))).....- ( -36.90, z-score = -4.06, R) >dp4.chrXL_group1e 6208347 101 + 12523060 -AAAUUAACUUCUGUUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGUUCUACGA- -................(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))).............- ( -28.60, z-score = -1.69, R) >droPer1.super_21 341693 101 + 1645244 -AAAUUUACUUCUGUUCGACCGUGACA----GGACUCGAACCUGCAAUCUUCGGAU-CCGAAGUCCGACGC-CUUAUCCA-UUAGGCCACACGGUCAGUUGUUCUACGA- -................(((((((.((----((.......))))......((((((-.....)))))).((-((......-..))))..))))))).............- ( -28.60, z-score = -1.69, R) >droMoj3.scaffold_6496 1573678 103 + 26866924 AACAUCAGGUGUGUUGCGAUUCUGCCA----GGAGUCGAACCUGGAAUCUUCUGAU-CCGUAGUCAGACGC-GUUAUCCA-UUGCGCCACAGGACCAAUGAAUCGGCCCA ....((((((......(((((((....----))))))).)))))).....((((((-.....)))))).((-((......-..))))....((.((........)).)). ( -28.90, z-score = -0.09, R) >droWil1.scaffold_180700 720069 91 + 6630534 -----------AUUUUCGAUCCUGCCA----GGAGUCGAACCUGGAAUCUUCUGAU-CCGUAGUCAGACGC-GUUAUCCA-UUGCGCCACAGGACCGGUUGAUCCACAG- -----------....(((.((((((((----((.......))))).....((((((-.....)))))).((-((......-..))))..))))).)))...........- ( -26.00, z-score = -1.12, R) >anoGam1.chr2R 11360139 98 - 62725911 -----------AGAUUUGUGUGUGAUAAACUGUGUGCGUGUUUGUGAUCUUCUUCUAUCGGAUGGUGAUAUGGCCGUCCAUUUGUUUUUUACAUUCAUUUGCUUUGAUA- -----------.....((.((((((.((((.(((..((.((((((.(((.(((......))).))).))).)))))..)))..)))).)))))).))............- ( -20.70, z-score = -1.15, R) >droVir3.scaffold_12875 4777428 91 - 20611582 -----------AACAACGAUCCUGCCA----GGAGUCGAACCUGGAAUCUUCUGAU-CCGUAGUCAGACGC-GUUAUCCA-UUGCGCCACAGGACCGACUGAUCGUCCA- -----------....((((((((....----))(((((..((((......((((((-.....)))))).((-((......-..))))..))))..)))))))))))...- ( -28.30, z-score = -1.78, R) >droGri2.scaffold_15245 18116430 88 + 18325388 --------AACGUCUUCGAUCCUGCCA----GGAGUCGAACCUGGAAUCUUCUGAU-CCGUAGUCAGACGC-GUUAUCCA-UUGCGCCACAGGACCGAUUGAU------- --------...(((.(((.((((((((----((.......))))).....((((((-.....)))))).((-((......-..))))..))))).)))..)))------- ( -28.20, z-score = -1.88, R) >consensus ___________AGGUUCGACCGUGACA____GGACUCGAACCUGCAAUCUUCGGAU_CCGAAGUCCGACGC_CUUAUCCA_UUAGGCCACACGGUCAGUUGAUUUACGA_ .................(.(((((..........................((((((......)))))).((.((.........))))..))))).).............. (-15.42 = -12.82 + -2.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:48 2011