
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,887,506 – 13,887,563 |
| Length | 57 |
| Max. P | 0.997813 |

| Location | 13,887,506 – 13,887,563 |
|---|---|
| Length | 57 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 76.18 |
| Shannon entropy | 0.47369 |
| G+C content | 0.47156 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
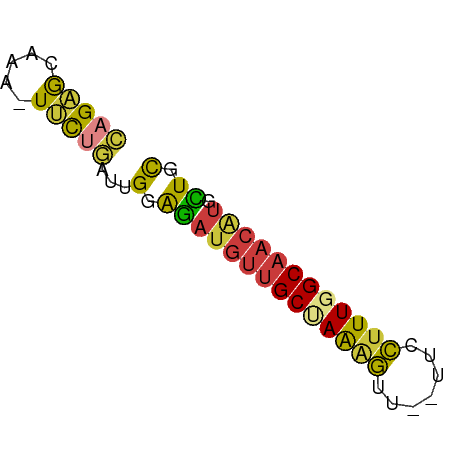
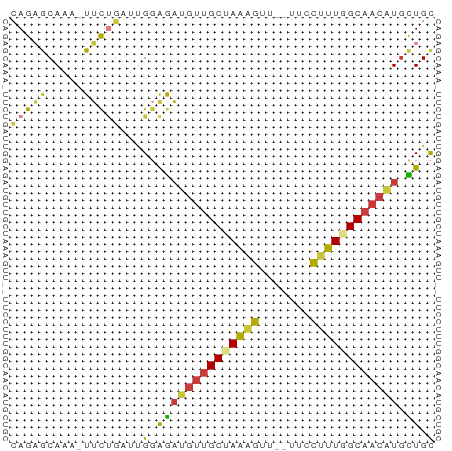
>dm3.chrX 13887506 57 + 22422827 CAGAACAAA-UUCUGAUUGGAGAUGUUGCUAAAGUUCCUUCCUUUGGCAACAUGCUGC (((((....-)))))...(.((((((((((((((.......)))))))))))).)).) ( -18.70, z-score = -3.24, R) >droSim1.chrX 10690153 55 + 17042790 CAGAGCAAA-UUCUGAUUGGAGAUGUUGCUAAAGUU--UUCCUUUGGCAACGUGCUGC ...(((...-((((....))))((((((((((((..--...))))))))))))))).. ( -18.40, z-score = -2.79, R) >droSec1.super_20 528518 54 + 1148123 CAGAGCAA--UUCUGAUUGGAGAUGUUGCUAAAGUU--UUCCUUUGGCAACGUGCUGC ...(((..--((((....))))((((((((((((..--...))))))))))))))).. ( -18.40, z-score = -2.81, R) >droYak2.chrX 8181400 56 + 21770863 CAGAGCAAAAUUCUGAUUGGAGAUGUUGCCAAAGUU--UUCCUUUGGCAACAUGCUGC ...(((....((((....))))((((((((((((..--...))))))))))))))).. ( -21.10, z-score = -3.57, R) >droEre2.scaffold_4690 5746531 55 - 18748788 CAGAGCAAA-UUCUGAUUGGAGAUGUUGCCAAAGUU--UUCCUUUGGCAACAUGCUGC ...(((...-((((....))))((((((((((((..--...))))))))))))))).. ( -21.10, z-score = -3.56, R) >droAna3.scaffold_13047 1416307 55 + 1816235 GAGCGCAUC-UUCGGGCCGUGAAUGUUGCGAAGCUU--CUCGUUUGGCAACAUUUUGU ..(((..((-....)).)))(((((((((.((((..--...)))).)))))))))... ( -17.70, z-score = -1.38, R) >droVir3.scaffold_12970 10146478 56 - 11907090 GAGUGCAGAGUGUCUGUUGGGGCGA--GCGAGCGCGCGCUCCUUUGGCAACAUGUUGC ....((((.((((.(((..(((.((--(((......))))))))..))))))).)))) ( -23.00, z-score = -1.16, R) >consensus CAGAGCAAA_UUCUGAUUGGAGAUGUUGCUAAAGUU__UUCCUUUGGCAACAUGCUGC (((((.....)))))...(.((((((((((((((.......)))))))))))).)).) (-12.96 = -12.63 + -0.34)

| Location | 13,887,506 – 13,887,563 |
|---|---|
| Length | 57 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 76.18 |
| Shannon entropy | 0.47369 |
| G+C content | 0.47156 |
| Mean single sequence MFE | -15.20 |
| Consensus MFE | -7.99 |
| Energy contribution | -8.11 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
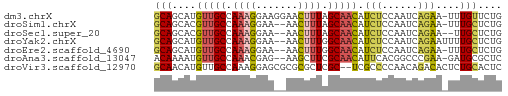
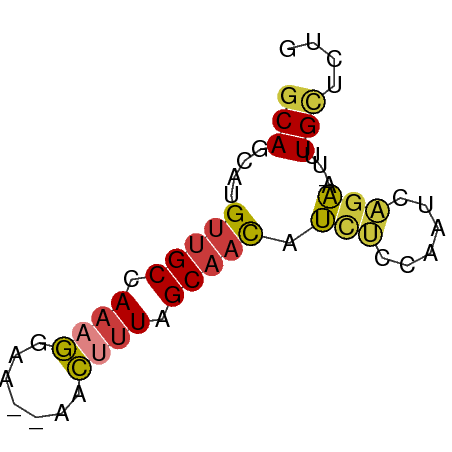
>dm3.chrX 13887506 57 - 22422827 GCAGCAUGUUGCCAAAGGAAGGAACUUUAGCAACAUCUCCAAUCAGAA-UUUGUUCUG (.((.(((((((.((((.......)))).))))))))).)...(((((-....))))) ( -15.00, z-score = -1.99, R) >droSim1.chrX 10690153 55 - 17042790 GCAGCACGUUGCCAAAGGAA--AACUUUAGCAACAUCUCCAAUCAGAA-UUUGCUCUG ..((((.(((((.((((...--..)))).))))).(((......))).-..))))... ( -13.10, z-score = -2.23, R) >droSec1.super_20 528518 54 - 1148123 GCAGCACGUUGCCAAAGGAA--AACUUUAGCAACAUCUCCAAUCAGAA--UUGCUCUG ..((((.(((((.((((...--..)))).))))).(((......))).--.))))... ( -13.10, z-score = -2.33, R) >droYak2.chrX 8181400 56 - 21770863 GCAGCAUGUUGCCAAAGGAA--AACUUUGGCAACAUCUCCAAUCAGAAUUUUGCUCUG (.((.((((((((((((...--..)))))))))))))).)...((((.......)))) ( -20.50, z-score = -4.19, R) >droEre2.scaffold_4690 5746531 55 + 18748788 GCAGCAUGUUGCCAAAGGAA--AACUUUGGCAACAUCUCCAAUCAGAA-UUUGCUCUG (.((.((((((((((((...--..)))))))))))))).)...((((.-.....)))) ( -21.00, z-score = -4.43, R) >droAna3.scaffold_13047 1416307 55 - 1816235 ACAAAAUGUUGCCAAACGAG--AAGCUUCGCAACAUUCACGGCCCGAA-GAUGCGCUC ....((((((((.((.(...--..).)).))))))))...(((.(...-...).))). ( -8.50, z-score = 0.75, R) >droVir3.scaffold_12970 10146478 56 + 11907090 GCAACAUGUUGCCAAAGGAGCGCGCGCUCGC--UCGCCCCAACAGACACUCUGCACUC ((((....))))....((.(((.((....))--.))).))..(((.....)))..... ( -15.20, z-score = -0.74, R) >consensus GCAGCAUGUUGCCAAAGGAA__AACUUUAGCAACAUCUCCAAUCAGAA_UUUGCUCUG (((....(((((.((((.......)))).))))).(((......)))....))).... ( -7.99 = -8.11 + 0.13)
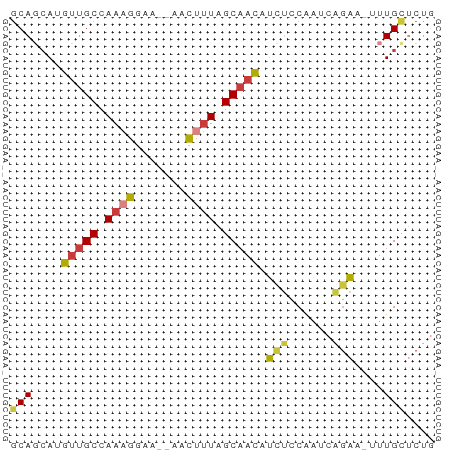
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:45 2011