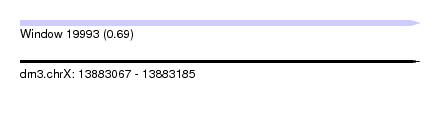
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,883,067 – 13,883,185 |
| Length | 118 |
| Max. P | 0.685020 |

| Location | 13,883,067 – 13,883,185 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 133 |
| Reading direction | forward |
| Mean pairwise identity | 65.15 |
| Shannon entropy | 0.65883 |
| G+C content | 0.54953 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
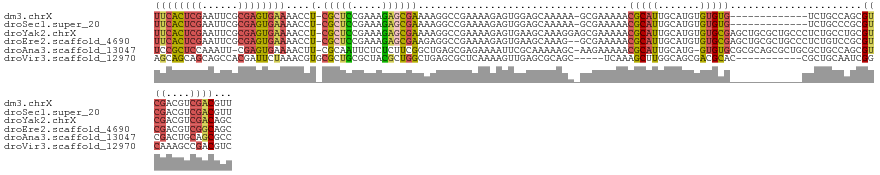
>dm3.chrX 13883067 118 + 22422827 UUCACUCGAAUUCGCGAGUGAAAACCU-CGCUCCGAAAGAGCGAAAAGGCCGAAAAGAGUGGAGCAAAAA-GCGAAAAACGCAUUGCAUGUGUGUG-------------UCUGCCAGCGUCGACGUCGACGUU ((((((((......))))))))....(-((((.(....))))))...(((.((..........((.....-)).....((((((.....)))))).-------------)).)))(((((((....))))))) ( -41.40, z-score = -2.44, R) >droSec1.super_20 524489 118 + 1148123 UUCACUCGAAUUCGCGAGUGAAAACCU-CGCUCCGAAAGAGCGAAAAGGCCGAAAAGAGUGGAGCAAAAA-GCGAAAAACGCAUUGCAUGUGUGUG-------------UCUGCCCGCGUCGACGUCGACGUU ((((((((......))))))))....(-((((.(....))))))...(((.((..........((.....-)).....((((((.....)))))).-------------)).))).((((((....)))))). ( -40.00, z-score = -1.98, R) >droYak2.chrX 8176914 132 + 21770863 UUCACUCGAAUUCGCGAGUGAAAACCU-CGCUCCGAAAGAGCGAAAAGGCCGAAAAGAGUGAAGCAAAGGAGCGAAAAACGCAUUGCAUGUGUGCGAGCUGCGCUGCCCUCUGCCUGCGUCGACGUCGACAGC ((((((((......))))))))....(-((((.(....))))))..((((.((....((((.(((......(((.....))).(((((....)))))))).))))....)).))))..((((....))))... ( -47.40, z-score = -1.70, R) >droEre2.scaffold_4690 5742148 130 - 18748788 UUCACUCGAAUUCGCGAGUGAAAACCU-CGCUCCGAAAGAGCGAAGAGGCCGAAAAGAGUGAAGCAAAG--GCGAAAAACGCAUUGCAUGUGUGCGAGCUGCGCUGCCCUCUGUCCGCGUCGACGUCGGCAGC ((((((((......))))))))..(.(-((((.(....)))))).)..(((((..((((.(.(((...(--((.....((((((...))))))....)))..))).).))))(((......))).)))))... ( -46.90, z-score = -1.01, R) >droAna3.scaffold_13047 1411517 129 + 1816235 UCCGCUCCAAAUU-CGAGUGAAAACUU-CGCAAUUCUCUCUUCGGCUGAGCGAGAAAAUUCGCAAAAAGC-AAGAAAAACGCAUUGCAUG-GUGUGCGCGCAGCGCUGCGCUGCCAGCGUCGACUGCAGCGCC ...((.(((....-.((((....))))-.(((((.(....(((.(((..(((((....)))))....)))-..)))....).))))).))-).))((((((((((..((((.....)))))).)))).)))). ( -39.70, z-score = 0.94, R) >droVir3.scaffold_12970 10140436 117 - 11907090 AGCAGCAGCAGCCACGAUUCUAAACGUGCGCUGCGCUACGCUGGCUGAGCGCUCAAAAGUUGAGCGCAGC-----UCAAAGCUUGGCAGCGACGCAC-----------CGCUGCAAUCGGCAAAGCCGACGUC .(((((.(((((((((........)))).)))))((..(((((.(...(((((((.....)))))))(((-----.....))).).)))))..))..-----------.)))))..(((((...))))).... ( -57.00, z-score = -2.87, R) >consensus UUCACUCGAAUUCGCGAGUGAAAACCU_CGCUCCGAAAGAGCGAAAAGGCCGAAAAGAGUGGAGCAAAGA_GCGAAAAACGCAUUGCAUGUGUGCG____________CUCUGCCAGCGUCGACGUCGACGUC ((((((((......))))))))......(((((.....)))))....................................(((((.......)))))......................((((....))))... (-15.92 = -16.40 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:42 2011