
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,872,201 – 13,872,291 |
| Length | 90 |
| Max. P | 0.865872 |

| Location | 13,872,201 – 13,872,291 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.16 |
| Shannon entropy | 0.49402 |
| G+C content | 0.48680 |
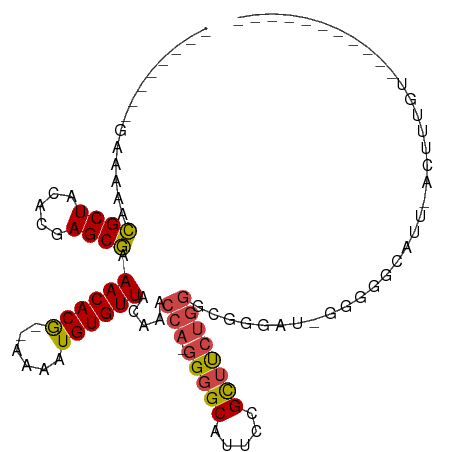
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.865872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
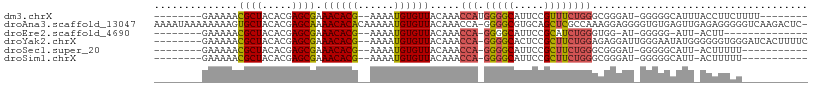
>dm3.chrX 13872201 90 + 22422827 --------GAAAAACGCUACACGAGCGAAACACG--AAAAUGUGUUACAAACCAUGGGGCAUUCCGUUUCUGGGCGGGAU-GGGGGCAUUUACCUUCUUUU-------- --------......((((.....)))).(((((.--.....))))).............(((((((((....))))))))-)(((.......)))......-------- ( -21.20, z-score = -0.18, R) >droAna3.scaffold_13047 1399470 107 + 1816235 AAAAUAAAAAAAAGUGCUACACGAGCAAAACACACAAAAAUGUGUUACAAACCA-GGGGCGUGCAGCUCGCCAAAGGAGGGGUGUGAGUUGAGAGGGGGUCAAGACUC- ..............((((.....)))).((((((......))))))(((..((.-..((((.......))))......))..)))(((((..((.....))..)))))- ( -22.80, z-score = -0.17, R) >droEre2.scaffold_4690 5731578 80 - 18748788 --------GAAAAACGCUACACGAGCGAAACACG--AAAAUGUGUUACAAACCA-GGGGCAUUCCGCAUCUGGGUGG-AU-GGGGG-AUU-ACUU-------------- --------......((((.....)))).(((((.--.....)))))........-(((..(((((.(((((....))-))-).)))-)).-.)))-------------- ( -21.10, z-score = -1.72, R) >droYak2.chrX 8166228 98 + 21770863 --------GAAAAACGCUACACGAGCGAAACACG--AAAAUGUGUUACAAACCA-GGGGCACUCCGCUUCUGGAGAGGAUUGGGAAUAUGGGGGGUGGGAUCACUUUUC --------......((((.....)))).(((((.--.....))))).....(((-(((((.....))))))))................(..(((((....)))))..) ( -26.60, z-score = -1.63, R) >droSec1.super_20 514052 85 + 1148123 --------GAAAAACGCUACACGAGCGAAACACG--AAAAUGUGUUACAAACCA-GGGGCAUUCCGCUUCUGGGCGGGAU-GGGGGCAUU-ACUUUUU----------- --------.......(((.(((..(((.(((((.--.....))))).)...(((-(((((.....))))))))))..).)-)..)))...-.......----------- ( -24.00, z-score = -2.00, R) >droSim1.chrX 10676064 85 + 17042790 --------GAAAAACGCUACACGAGCGAAACACG--AAAAUGUGUUACAAACCA-GGGGCAUUCCGCUUCUGGGCGGGAU-GGGGGCAUU-ACUUUUU----------- --------.......(((.(((..(((.(((((.--.....))))).)...(((-(((((.....))))))))))..).)-)..)))...-.......----------- ( -24.00, z-score = -2.00, R) >consensus ________GAAAAACGCUACACGAGCGAAACACG__AAAAUGUGUUACAAACCA_GGGGCAUUCCGCUUCUGGGCGGGAU_GGGGGCAUU_ACUUUGU___________ ..............((((.....)))).((((((......)))))).....(((.(((((.....)))))))).................................... (-14.57 = -14.72 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:41 2011