
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,870,748 – 13,870,892 |
| Length | 144 |
| Max. P | 0.639677 |

| Location | 13,870,748 – 13,870,892 |
|---|---|
| Length | 144 |
| Sequences | 7 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.32810 |
| G+C content | 0.56294 |
| Mean single sequence MFE | -51.11 |
| Consensus MFE | -24.93 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13870748 144 + 22422827 CCCUUUUCGGCACUGCCAUGUU------GUCCUCGGUUAUGUC-CGAUGGACGCGAUAACGGCAAAAACGACGCCGGCUG--------CUCUCCGCCUGU------GUCAAC--GUUGUCG-UCAUCGUCGGGGGCUAAUAAACUCUCCCGUUUAGGCUGUCGCAUAC .......(((((..(((.....------((((((((......)-))).))))(((((.(((((((....(((((.(((..--------......))).))------)))...--.))))))-).)))))((((((..........))))))....))))))))..... ( -55.60, z-score = -3.42, R) >droSim1.chrX 10674674 144 + 17042790 CCCUUUUCGGCACUGCCAUGUU------GUCCUCGGUUAUGUC-CGAUGGACGCGAUAACAGCAAAAACGACGCCGGCUG--------CUCUCCGCCUGU------GUCAAC--GUUGUCG-UCAUCGUCGGGGGCUAAUAAACUCUCCCGUUUAGGUUGUCGCAUAC ........(((...))).....------((((((((......)-))).))))((((((((.((((....(((((.(((..--------......))).))------)))...--.))))..-.......((((((..........)))))).....)))))))).... ( -49.30, z-score = -2.08, R) >droSec1.super_20 512605 144 + 1148123 CCCUUUUCGGCACUGCCAUGUU------GUCCUCGGUUAAGUC-CGAUGGACGCGAUAACAGCAAAAACGACGCCGGCUG--------CUCUCCGCCUGU------GUCAAC--GUCGUCG-UCAUGGUCGGGGGCUAAUAAACUCUCCCGUUUAGGUUGUCGCAUAC ........(((...))).....------((((((((......)-))).))))(((((((((((......(((((.(((..--------......))).))------)))..(--.(((.(.-.....).))).))))..(((((......))))).)))))))).... ( -49.10, z-score = -1.63, R) >droYak2.chrX 8164838 144 + 21770863 CCCUUUUCGGCACUGCCAUGCU------GUCCUCGGUUAUGUC-CGAUGGACGCGAUAACGGCAAAAACGACGCCGGCUG--------CUCUCCGCCUGU------GUCAAC--GUUGUCG-UCAUCGUCGGGGGCUAAUAAACUCUCCCGUUUAGGUUGUCCCAUAC .......((((........((.------((((((((......)-))).))))))(((.(((((((....(((((.(((..--------......))).))------)))...--.))))))-).)))))))(((((...(((((......)))))....))))).... ( -55.40, z-score = -3.68, R) >droEre2.scaffold_4690 5730041 144 - 18748788 CCCUUUUCGGCACUGCCAUGUU------GUCCUCGGUUAUGUC-CGAUGGACGCGAUAACGGCAAAAACGACGCCGGCUG--------CUCUCCGCCUGU------GUCAAC--GUUGUCG-UCAUCGUCGGGGGCUAAUAAACUCUCCCGUUUAGGUUGUCCCAUAC ........((.((.(((.....------((((((((......)-))).))))(((((.(((((((....(((((.(((..--------......))).))------)))...--.))))))-).)))))((((((..........))))))....))).)).)).... ( -54.30, z-score = -3.39, R) >dp4.chrXL_group1e 6168399 154 - 12523060 -CCCGUUCGACGCUGCUGGGUUCCUCCUGUCCACGCUGAUGUCGCGAUGAACGCGAUAACGGCACAAGCGACGCCGGCUGUCCUGUCCCUCUCGCUCUGUCCCACUGUCAACUUGUUGUUGGCUGUCGUCGGGGGCUAAUAAACUCUCCCAUUUA------------- -((((..((((.....((((..............((((.(((((((.....))))))).))))((((((((....(((......)))....))))).)))))))..((((((.....)))))).)))).))))......................------------- ( -50.40, z-score = -1.33, R) >droPer1.super_21 303151 154 - 1645244 CCCCGUUCGACGCUGCUGGGUUCCUCCUGUCCACGCUGAUGUCGCGAUGAACGCGAUAACGGCAAAACCGACGCCGGCUGUCCUGUCUCUCUCGCUCUGUCCCACUGUCAACUUGUUGUUG-CUGUCGUCGGGGGCUAAUAAACUCUCCCAUUUA------------- (((((..((((((.((((((((......(....)((((.(((((((.....))))))).))))......))).))))).)).........................(.((((.....))))-).)))).))))).....................------------- ( -43.70, z-score = -0.71, R) >consensus CCCUUUUCGGCACUGCCAUGUU______GUCCUCGGUUAUGUC_CGAUGGACGCGAUAACGGCAAAAACGACGCCGGCUG________CUCUCCGCCUGU______GUCAAC__GUUGUCG_UCAUCGUCGGGGGCUAAUAAACUCUCCCGUUUAGGUUGUCGCAUAC ((((....((((..(((.................)))..)))).((((((...((((((((((.........)))(((................))).................)))))))....))))))))))....(((((......)))))............. (-24.93 = -25.26 + 0.33)

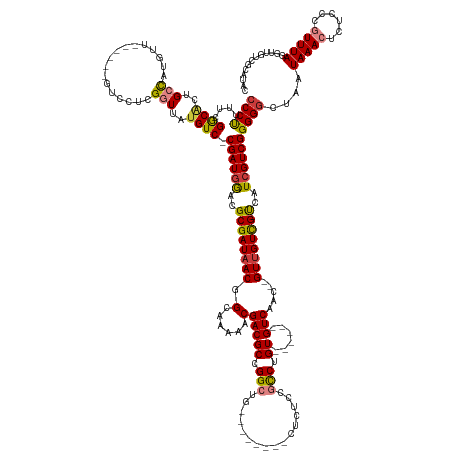
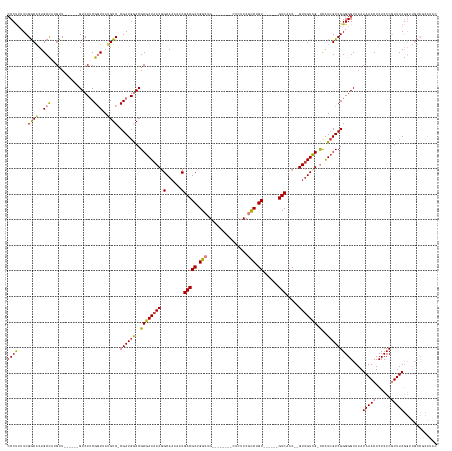
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:40 2011