| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,369,274 – 11,369,364 |
| Length | 90 |
| Max. P | 0.868410 |

| Location | 11,369,274 – 11,369,364 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.65449 |
| G+C content | 0.55245 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
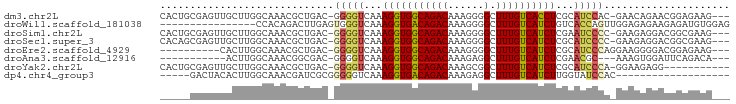
>dm3.chr2L 11369274 90 - 23011544 CACUGCGAGUUGCUUGGCAAACGCUGAC-GGGGUCAAAGGUGGCAGACAAAGGGGCUUUGUCACCUCGCAUCCAC-GAACAGAACGGAGAAG--- ..((.((..(((.(..((....))..)(-(((((...(((((((((((......).))))))))))...)))).)-)..)))..)).))...--- ( -26.50, z-score = -0.21, R) >droWil1.scaffold_181038 275842 79 + 637489 ----------------CCACAGACUUGAGUGGGUCAAAGGUGACAGACAAAGGGGCUUUGUCAUCUGUCACCAGUUGGAGAGAAGAGAUGUGGAG ----------------((((((((((....)))))...((((((((((((((...)))))))...)))))))................))))).. ( -28.40, z-score = -2.73, R) >droSim1.chr2L 11175370 90 - 22036055 CACUGCGAGUUGCUUGGCAAACGCUGAC-GGGGUCAAAGGUGGCAGACAAAGGGGCUUUGUCAUCUCGAAUCCCC-GAAGAGGACGGCGAAG--- ...(((..(((.((((((....)))..(-((((....(((((((((((......).)))))))))).....))))-))))..))).)))...--- ( -27.60, z-score = 0.07, R) >droSec1.super_3 6763797 90 - 7220098 CACAGCGAGUUGCUUGGCAAACGCUGAC-GGGGUCAAAGGUGGCAGACAAAGGGGCUUUGUCAUCUCGCAUCCCC-GAAGAGGACGGCGAAG--- ..(((((..((((...)))).))))).(-((((....(((((((((((......).)))))))))).....))))-)...............--- ( -30.40, z-score = -0.77, R) >droEre2.scaffold_4929 12566632 81 + 26641161 ----------CACUUGGCAAACGCUGAC-GGGGUCAAAGGUGGCAGACAAAGGGGCUUUGUCAUCUCGCAUCCCAGGAAGGGGACGGAGAAG--- ----------((((((((...((....)-)..))))..))))...(((((((...))))))).((((.(.((((......)))).)))))..--- ( -25.50, z-score = -1.14, R) >droAna3.scaffold_12916 12080490 77 + 16180835 -----------ACUUGGCAAACGGCGAC-GGGGUCAAAGGUGGCAGACAAAGAGGCUUUGUCAUCUCGAACGC---AAAGUGGAUUCAGACA--- -----------..((((...((.(((.(-(((((((....)))).(((((((...)))))))..))))..)))---...))....))))...--- ( -18.80, z-score = -0.14, R) >droYak2.chr2L 7765400 82 - 22324452 CACUGCGAGUUGCUUGGCAAACGCUGAC-GGGGUCAAAGGUGGCAGACAAAGCGGCUUUGUCAUCUCGCAUCCCA-GGAAGAGG----------- ...((((((.....(((((((.((((..-.(.((((....))))...)....))))))))))).))))))((...-.)).....----------- ( -24.60, z-score = 0.19, R) >dp4.chr4_group3 6812504 71 - 11692001 -----GACUACACUUGGCAAACGAUCGCGGGGGUCAAAGGUGACAGACAAAGAGGCUUUGUCAUCUUGGUAUCCAC------------------- -----...........((........))(((..((((.((((((((((......).)))))))))))))..)))..------------------- ( -17.70, z-score = -0.77, R) >consensus _____CGAGUUGCUUGGCAAACGCUGAC_GGGGUCAAAGGUGGCAGACAAAGGGGCUUUGUCAUCUCGCAUCCCA_GAAGAGGACGGAGAAG___ .............................(((((...(((((((((((......).))))))))))...)))))..................... (-14.48 = -14.52 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:12 2011