| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,868,373 – 13,868,502 |
| Length | 129 |
| Max. P | 0.765654 |

| Location | 13,868,373 – 13,868,502 |
|---|---|
| Length | 129 |
| Sequences | 11 |
| Columns | 150 |
| Reading direction | forward |
| Mean pairwise identity | 69.61 |
| Shannon entropy | 0.62039 |
| G+C content | 0.49305 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.31 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13868373 129 + 22422827 CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGCAA-------------CCGAGUCGCCGAGCG-AGUCGGCGAAACUACAAAGGACCUCAAAGCCC--AAGACGAUGACGUCAAAGGGG----- .............(((......((((....))))(((((....))))).........))).-------------..((((((((((...-..)))))))..((....))...)))....(((--..((((....))))...))).----- ( -36.60, z-score = -2.05, R) >droSim1.chrX 10672368 129 + 17042790 CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGCAA-------------CCGAGUCGCCGAGCG-AGUCGGCGAAACUACAAAGGACCUCAAAGCCC--AAGACGAUGACGUCAAAGGGG----- .............(((......((((....))))(((((....))))).........))).-------------..((((((((((...-..)))))))..((....))...)))....(((--..((((....))))...))).----- ( -36.60, z-score = -2.05, R) >droSec1.super_20 510292 129 + 1148123 CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGCAA-------------CCGAGUCGCCGAGCG-AGUCGGCGAAACUACAAAGGACCUCAAAGCCC--AAGACGAUGACGUCAAAGGGG----- .............(((......((((....))))(((((....))))).........))).-------------..((((((((((...-..)))))))..((....))...)))....(((--..((((....))))...))).----- ( -36.60, z-score = -2.05, R) >droYak2.chrX 8161830 129 + 21770863 CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGCAG-------------CCGAGUCGUCGAGCG-AGUCGGCAAAACUACAAAGGACCUCAAAGCCC--AAGACGAUGACGUCAAAGGGG----- .(((.........(((......((((....))))(((((....))))).........)))(-------------((((.(((.....))-).)))))...........)))........(((--..((((....))))...))).----- ( -39.30, z-score = -2.92, R) >droEre2.scaffold_4690 5727832 129 - 18748788 CUCUACGAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAUGCAA-------------CCGAGUCGCCGAGCG-AGUCGGCGAAACUACAAAGGACCUCAAAGCCC--AAGACGAUGACGUCAAAGGGG----- .............(((((....((((....))))(((((....))))).......))))).-------------..((((((((((...-..)))))))..((....))...)))....(((--..((((....))))...))).----- ( -38.40, z-score = -2.20, R) >droAna3.scaffold_13047 1396301 122 + 1816235 CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGGCAAAAAG---------------------GAUUCAAGUCGUCGGCGAA-CUACAAAGGACCUCAAAAGCC-CAAGACGAUGAUGCCAAACGAA----- .............(((......((((....))))(((((....)))))....)))....(---------------------(......(((((((((...-..................)))-...))))))....)).......----- ( -27.50, z-score = -1.14, R) >droVir3.scaffold_12970 10120520 106 - 11907090 CCCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCAACAAGUAGGCAAUACCCUGCGAG----------------------GAUGACGCGUCGGUGCAACUACAAAGA----GAGACAGC-GGCU-CAGGGG---------------- ((((........((((......))))..(((((((((.....(((((......)))))((.----------------------(......).))))))).))))......----(((.....-..))-))))).---------------- ( -29.30, z-score = -0.98, R) >droGri2.scaffold_15081 2755585 113 - 4274704 CCCUACAAAAUUAUGCAUAAACGCAUUUUACUGCGCCUCCAAGUAGGCAAUGCCCACUGAG----------------------GAUGUCGCCUCGGCGGAACUACAAAGGCCAGGGAACAGC-GACGGCAAACGGG-------------- (((.........((((......))))........((((((.(((.(((...))).)))..)----------------------)).(((((((((((............))).)))....))-))))))....)))-------------- ( -32.20, z-score = -0.50, R) >droWil1.scaffold_181096 8460251 113 + 12416693 ----ACAAAAUUAUGCAUAAACGCACUUUAGUGCGCCUACAAGUAGGCAAUGUCAAAA-GG-------------AAGCGAAGGCACAAA-AAACUACAAAGCGUUAA---CUCAAAGAGGGC--AAGACGAUGUGGG------------- ----..........((((....((((....))))(((((....))))).)))).....-..-------------..((....)).....-...(((((...((((..---(((.....))).--..)))).))))).------------- ( -28.30, z-score = -2.41, R) >droPer1.super_21 301031 145 - 1645244 ---UGCAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCAAAAGGACCCCGCGCACACACACACUCACACACACGGAGUCGGCAAAACUACAAAGGACCUCAAAGCCCACAAGACG--GAUGCCCACGGUGGCGGA ---..........(((((....((((....))))(((((....))))).)))))........((((.(((......((((.(......))))).((((...((....))..((((...........)).)--).))))....))))))). ( -35.90, z-score = -0.32, R) >dp4.chrXL_group1e 6166396 145 - 12523060 ---UGCAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGGACCCCGCACACACACACACACAGACACACGGAGUCGGCAAAACUACAAAGGACCUCAAAGCCCACAAGACG--GAUGCCCACGGUGGCGGA ---.(((.....((((......)))).....)))(((((....)))))..(((((..((((.(((......................))).)))((((...((....))..((((...........)).)--).))))..)..))))).. ( -34.65, z-score = -0.29, R) >consensus CUCUACAAAAUUAUGCAUAAACGCAUUUUAGUGCGCCUACAAGUAGGCAAUGCCAAAGCAA_____________CCGAGUCGCCAAGCG_AGUCGGCGAAACUACAAAGGACCUCAAAGCCC__AAGACGAUGACGUCAAAGGGG_____ .............(((......((((....))))(((((....)))))...............................................))).................................................... (-13.33 = -13.31 + -0.02)

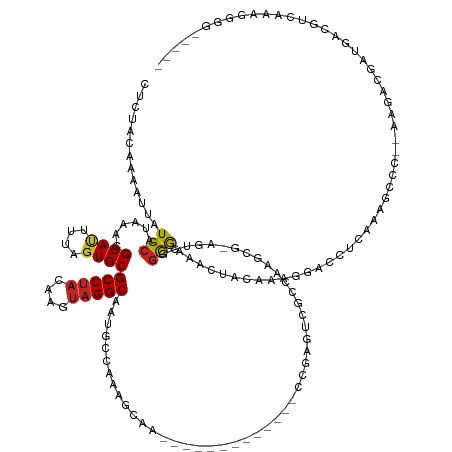
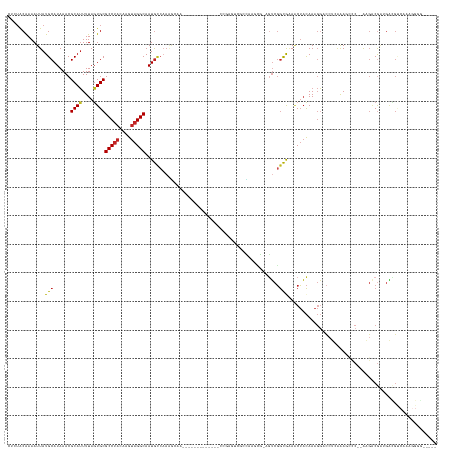
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:39 2011