
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,866,146 – 13,866,206 |
| Length | 60 |
| Max. P | 0.646656 |

| Location | 13,866,146 – 13,866,206 |
|---|---|
| Length | 60 |
| Sequences | 7 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.44541 |
| G+C content | 0.49524 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
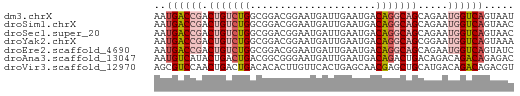
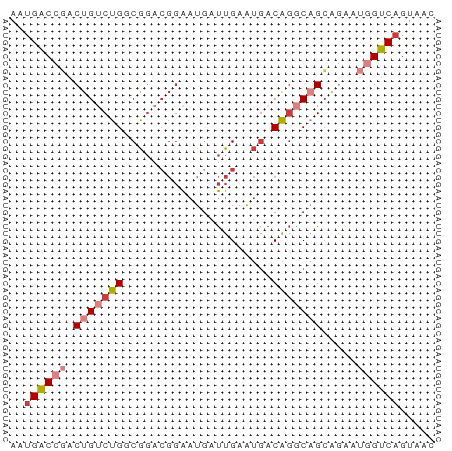
>dm3.chrX 13866146 60 + 22422827 AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCAGAAUGGUCAGUAAU ..((((((.(((((((.((..(((.....)))..)).))))))).....))))))..... ( -15.90, z-score = -1.74, R) >droSim1.chrX 10670166 60 + 17042790 AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCAGAAUGGUCAGUAAC ..((((((.(((((((.((..(((.....)))..)).))))))).....))))))..... ( -15.90, z-score = -1.77, R) >droSec1.super_20 508128 60 + 1148123 AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCAGAAUGGUCAGUAAC ..((((((.(((((((.((..(((.....)))..)).))))))).....))))))..... ( -15.90, z-score = -1.77, R) >droYak2.chrX 8159543 60 + 21770863 AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCGGAAUGGUCAGUAAA ..((((((.(((((((.((..(((.....)))..)).))))))).....))))))..... ( -15.90, z-score = -1.59, R) >droEre2.scaffold_4690 5725501 60 - 18748788 AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCAGAAUGGUCAGUAUC ..((((((.(((((((.((..(((.....)))..)).))))))).....))))))..... ( -15.90, z-score = -1.31, R) >droAna3.scaffold_13047 1394416 60 + 1816235 AAUGUCAUACUGACUGACGGCGGGAAUGAUUGAAUGACAGACUGACAGACAGACAGAGAC .........(((.(((.((((.(..((......))..).).))).))).)))........ ( -9.10, z-score = -0.92, R) >droVir3.scaffold_12970 1788157 60 + 11907090 AGCGUCCAACUGACUGACACACUUGUUCACUGAGCAACGAGCUGCAUGACAGACAGACGU .(((((...(((.(((.((..((((((........)))))).)))).).)))...))))) ( -12.20, z-score = 0.49, R) >consensus AAUGACCGACUGUCUGGCGGACGGAAUGAUUGAAUGACAGGCAGCAGAAUGGUCAGUAAC ..((((((.(((((((.....................))))))).....))))))..... ( -8.93 = -9.83 + 0.90)

| Location | 13,866,146 – 13,866,206 |
|---|---|
| Length | 60 |
| Sequences | 7 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.44541 |
| G+C content | 0.49524 |
| Mean single sequence MFE | -11.82 |
| Consensus MFE | -7.27 |
| Energy contribution | -7.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
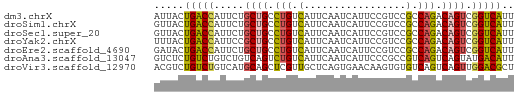
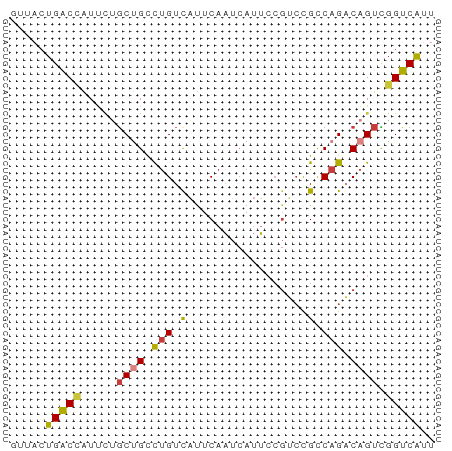
>dm3.chrX 13866146 60 - 22422827 AUUACUGACCAUUCUGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((...((((((.(.(.(.............).).).))).)))..))))).. ( -11.22, z-score = -1.35, R) >droSim1.chrX 10670166 60 - 17042790 GUUACUGACCAUUCUGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((...((((((.(.(.(.............).).).))).)))..))))).. ( -11.22, z-score = -1.09, R) >droSec1.super_20 508128 60 - 1148123 GUUACUGACCAUUCUGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((...((((((.(.(.(.............).).).))).)))..))))).. ( -11.22, z-score = -1.09, R) >droYak2.chrX 8159543 60 - 21770863 UUUACUGACCAUUCCGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((.....((((.(((.(.................).))).)))).))))).. ( -10.03, z-score = -1.07, R) >droEre2.scaffold_4690 5725501 60 + 18748788 GAUACUGACCAUUCUGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((...((((((.(.(.(.............).).).))).)))..))))).. ( -11.22, z-score = -0.70, R) >droAna3.scaffold_13047 1394416 60 - 1816235 GUCUCUGUCUGUCUGUCAGUCUGUCAUUCAAUCAUUCCCGCCGUCAGUCAGUAUGACAUU (((.(((.(((.(.((..(..((.........))..)..)).).))).)))...)))... ( -11.40, z-score = -2.72, R) >droVir3.scaffold_12970 1788157 60 - 11907090 ACGUCUGUCUGUCAUGCAGCUCGUUGCUCAGUGAACAAGUGUGUCAGUCAGUUGGACGCU .((.(((.(((.((((((((.....)))..(....)..))))).))).))).))...... ( -16.40, z-score = 0.01, R) >consensus GUUACUGACCAUUCUGCUGCCUGUCAUUCAAUCAUUCCGUCCGCCAGACAGUCGGUCAUU .....(((((.....((((.(((.(.................).))).)))).))))).. ( -7.27 = -7.12 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:37 2011