| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,828,825 – 13,828,921 |
| Length | 96 |
| Max. P | 0.987321 |

| Location | 13,828,825 – 13,828,921 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Shannon entropy | 0.54107 |
| G+C content | 0.46311 |
| Mean single sequence MFE | -19.71 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
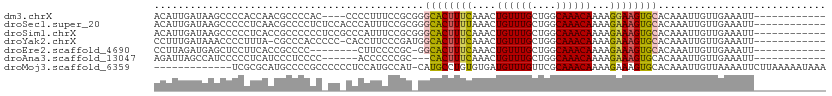
>dm3.chrX 13828825 96 + 22422827 ACAUUGAUAAGCCCCACCAACGCCCCAC----CCCCUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGGAAGUGCACAAAUUGUUGAAAUU------------ ...(..((((..........(((.....----.........))).((((((((....((((((....))))))...)))))))).....))))..)....------------ ( -21.24, z-score = -0.52, R) >droSec1.super_20 474718 100 + 1148123 ACAUUGAUAAGCCCCCUCAACGCCCCUCUCCACCCAUUUCCGCGGGCACUUUUAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU------------ ................(((((............((........))((((((((....((((((....))))))...)))))))).......)))))....------------ ( -21.40, z-score = -1.45, R) >droSim1.chrX 10642529 100 + 17042790 ACAUUGAUAAGCCCCCUCACCGCCCCCCUCCGCCCAUUUCCGCGGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU------------ ...(..((((...................((((........))))((((((((....((((((....))))))...)))))))).....))))..)....------------ ( -25.90, z-score = -2.69, R) >droYak2.chrX 8123443 98 + 21770863 CCUUUGAUAAACCCCUUUA-CGCCCACCCCC-CACCUUCCCGAUGGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU------------ ..((..((((.........-...........-((.(.....).))((((((((....((((((....))))))...)))))))).....))))..))...------------ ( -19.20, z-score = -2.16, R) >droEre2.scaffold_4690 5690776 91 - 18748788 CCUUAGAUGAGCUCCUUCACCGCCCC--------CUUCCCCGC-GGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU------------ ...............((((.(((...--------.......))-)((((((((....((((((....))))))...)))))))).........))))...------------ ( -20.50, z-score = -1.52, R) >droAna3.scaffold_13047 1356816 91 + 1816235 AGAUUAGCCAUCCCCCUCAUCCCUCCCC------ACCCCCCGC---CACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU------------ ............................------.......((---.((((((....((((((....))))))...))))))))................------------ ( -13.80, z-score = -1.54, R) >droMoj3.scaffold_6359 1483258 98 - 4525533 -------------UCGCGCAUGCCCCGCCCCCCUCCAUGCCAU-CAUGCCUGUGUGAUGUUUGUUCGCAAACAAAAGAAAGUGCACAAAUUGUUAAAAUUCUUAAAAAUAAA -------------(((((((.((...((..........))...-...)).)))))))((((((....)))))).(((((...(((.....))).....)))))......... ( -15.90, z-score = -1.08, R) >consensus ACAUUGAUAAGCCCCCUCAACGCCCCCC_CC_CCCCUUCCCGC_GGCACUUUCAAACUGUUUGCUGGCAAACAAAAGAAAGUGCACAAAUUGUUGAAAUU____________ .............................................((((((((....((((((....))))))...))))))))............................ (-14.18 = -14.20 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:35 2011