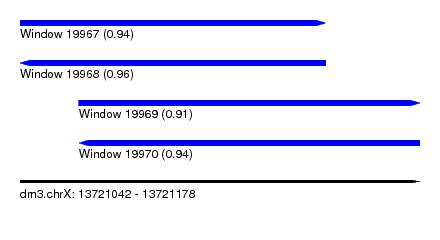
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,721,042 – 13,721,178 |
| Length | 136 |
| Max. P | 0.957109 |

| Location | 13,721,042 – 13,721,146 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.55 |
| Shannon entropy | 0.72677 |
| G+C content | 0.49629 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -13.16 |
| Energy contribution | -12.51 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
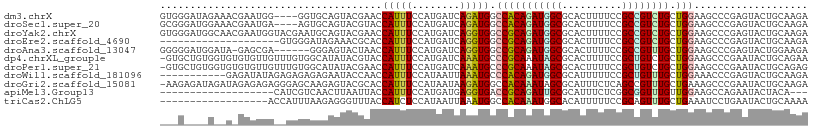
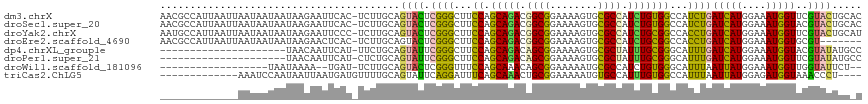
>dm3.chrX 13721042 104 + 22422827 GUGGGAUAGAAACGAAUGG----GGUGCAGUACGAACCAUUUCCAUGAUCAGAUGGCCACAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA ...................----..((((((((...((((((........))))))((((((((((((........))))))))).)))........))))))))... ( -35.70, z-score = -1.46, R) >droSec1.super_20 368235 104 + 1148123 GCGGGAUGGAAACGAAUGA----AGUGCAGUACGUACCAUUUCCAUGAUCAGAUGGCCACAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA ......((....)).....----..((((((((...((((((........))))))((((((((((((........))))))))).)))........))))))))... ( -37.50, z-score = -2.28, R) >droYak2.chrX 8012205 108 + 21770863 GUGGGAUGGCAACGAAUGGUACGAAUGCAGUACGAACCAUUUCCAUGAUCAGGUGGCCGCAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA ......((....))...........((((((((....(((....))).((.(((..((((((((((((........)))))))))).))..))).))))))))))... ( -41.30, z-score = -2.04, R) >droEre2.scaffold_4690 5583708 88 - 18748788 --------------------GUGGGAUAGAAACGCACCAUUUCCAUGAUCAGGUGGCCGCAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA --------------------((((((..............))))))..((.(((..((((((((((((........)))))))))).))..))).))........... ( -31.64, z-score = -1.41, R) >droAna3.scaffold_13047 1051073 101 + 1816235 GGGGGAUGGAUA-GAGCGA------GGGAGUACUAACCAUUUCCAUGAUCAGGUGGCCGCAGAUGGCGCACUUUUCCGCCGUUUGCUGGAAGCCCGAGUACUGGAAGA .((..((((.((-(.((..------....)).))).))))..))....((.(((..((((((((((((........)))))))))).))..))).))........... ( -40.50, z-score = -2.53, R) >dp4.chrXL_group1e 6020961 107 - 12523060 -GUGCUGUGGUGUGUGUUGUUUGUGGCAUAUACGUACCAUUUCCAUGAUCAAAUGCCCGCAAAUAGCGCACUUUUCCGCUGUCUGCUGGAAGCCCGAAUACUGCAGAA -...(((..((((((((((((((((((((.......................))).))))))))))))))).(((((((.....)).))))).......))..))).. ( -35.60, z-score = -2.32, R) >droPer1.super_21 134432 107 - 1645244 -GUGCUGUGGUGUGUGUUGUUUGUGGCAUAUACGAACCAUUUCCAUGAUCAAAUGCCCGCAAAUAGCGCACUUUUCCGCUGUCUGCUGGAAGCCCGAAUACUGCAGAG -...(((..((((((((((((((((((((....((..(((....))).))..))).))))))))))))))).(((((((.....)).))))).......))..))).. ( -35.90, z-score = -2.48, R) >droWil1.scaffold_181096 2221021 97 - 12416693 -----------GAGAUAUAGAGAGAGAGAAUACCAACCAUUUCCAUAAUUAAAUGCCCACAGAUGGCGCAUUUUUCCGCUGUUUGCUGGAAACCCGAGUACUGCAAGA -----------............................((((((.....(((((((((....))).))))))....((.....))))))))................ ( -14.40, z-score = 0.36, R) >droGri2.scaffold_15081 2590925 107 - 4274704 -AAGAGAUAGAUAGAGAGAGGGAGCAAGAGUACGCACCAUUUCCAUAAUAAGAUGGCCACAAAUAGCGCAUUUCUCAGCCGUUUGCUGAAAGCCCGAAUACUGCAAGA -........(.(((.....(((.....((((.(((.......((((......)))).........))).)))).(((((.....)))))...))).....)))).... ( -16.79, z-score = 1.13, R) >apiMel3.Group13 4637752 86 - 8929068 -------------------CAUCGUCAACUUAAUUACCAUUUCCAUGAUGAGGUGACCGCAGAUUGCGCAUUUCUCGGCGGUUUGUUGGAAGCCAGAAUACUACA--- -------------------..((((((....(((....)))....))))))(((..(((((((((((..........)))))))).)))..)))...........--- ( -19.60, z-score = -0.32, R) >triCas2.ChLG5 1206362 90 + 18847211 ------------------ACCAUUUAAGAGGGUUUACCAUCUCCAUAAUUAAAUGGCCACAAAUGGCACAUUUUUCCGCAGUUUGCUGAAAUCCUGAAUACUGCAAAA ------------------..(((((((...((..........))....)))))))((((....))))..........(((((...(.........)...))))).... ( -17.40, z-score = -0.71, R) >consensus ___G____G__A_GAGUGA__UGGGGAAAGUACGAACCAUUUCCAUGAUCAGAUGGCCGCAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA .....................................(((((........))))).(((((((((((..........)))))))).)))................... (-13.16 = -12.51 + -0.65)
| Location | 13,721,042 – 13,721,146 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.55 |
| Shannon entropy | 0.72677 |
| G+C content | 0.49629 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -15.09 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13721042 104 - 22422827 UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGUGGCCAUCUGAUCAUGGAAAUGGUUCGUACUGCACC----CCAUUCGUUUCUAUCCCAC ...((((((((...((((....(((((.((((........)))).)))))))))....((((((....)))))).))))))))..----................... ( -35.80, z-score = -2.16, R) >droSec1.super_20 368235 104 - 1148123 UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGUGGCCAUCUGAUCAUGGAAAUGGUACGUACUGCACU----UCAUUCGUUUCCAUCCCGC ...((((((((...((((....(((((.((((........)))).))))))))).....(((((....)))))..))))))))..----................... ( -35.30, z-score = -1.80, R) >droYak2.chrX 8012205 108 - 21770863 UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCACCUGAUCAUGGAAAUGGUUCGUACUGCAUUCGUACCAUUCGUUGCCAUCCCAC ...(((((((((((((...((.(((((.((((........)))).)))))))...)))))((((....))))...))))))))......................... ( -38.10, z-score = -1.96, R) >droEre2.scaffold_4690 5583708 88 + 18748788 UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCACCUGAUCAUGGAAAUGGUGCGUUUCUAUCCCAC-------------------- .......(((((((((...((.(((((.((((........)))).)))))))...))))).(((....))))))).............-------------------- ( -29.20, z-score = -0.76, R) >droAna3.scaffold_13047 1051073 101 - 1816235 UCUUCCAGUACUCGGGCUUCCAGCAAACGGCGGAAAAGUGCGCCAUCUGCGGCCACCUGAUCAUGGAAAUGGUUAGUACUCCC------UCGCUC-UAUCCAUCCCCC ..(((((....(((((...((.(((...((((........))))...)))))...)))))...)))))((((.(((..(....------..)..)-)).))))..... ( -27.50, z-score = -0.55, R) >dp4.chrXL_group1e 6020961 107 + 12523060 UUCUGCAGUAUUCGGGCUUCCAGCAGACAGCGGAAAAGUGCGCUAUUUGCGGGCAUUUGAUCAUGGAAAUGGUACGUAUAUGCCACAAACAACACACACCACAGCAC- ((((((.((......((.....))..)).))))))..((((((.....)).(((((.(((((((....))))).))...)))))...................))))- ( -27.60, z-score = -0.64, R) >droPer1.super_21 134432 107 + 1645244 CUCUGCAGUAUUCGGGCUUCCAGCAGACAGCGGAAAAGUGCGCUAUUUGCGGGCAUUUGAUCAUGGAAAUGGUUCGUAUAUGCCACAAACAACACACACCACAGCAC- ...(((.......((....)).(((((.((((........)))).))))).(((((.(((((((....)))).)))...)))))...................))).- ( -27.00, z-score = -0.24, R) >droWil1.scaffold_181096 2221021 97 + 12416693 UCUUGCAGUACUCGGGUUUCCAGCAAACAGCGGAAAAAUGCGCCAUCUGUGGGCAUUUAAUUAUGGAAAUGGUUGGUAUUCUCUCUCUCUAUAUCUC----------- ......((((((...(((((((((.....))....((((((.(((....))))))))).....)))))))....)))))).................----------- ( -20.40, z-score = -0.24, R) >droGri2.scaffold_15081 2590925 107 + 4274704 UCUUGCAGUAUUCGGGCUUUCAGCAAACGGCUGAGAAAUGCGCUAUUUGUGGCCAUCUUAUUAUGGAAAUGGUGCGUACUCUUGCUCCCUCUCUCUAUCUAUCUCUU- .............(((((((((((.....))))))).(((((((((((....((((......)))))))))))))))......))))....................- ( -26.40, z-score = -1.12, R) >apiMel3.Group13 4637752 86 + 8929068 ---UGUAGUAUUCUGGCUUCCAACAAACCGCCGAGAAAUGCGCAAUCUGCGGUCACCUCAUCAUGGAAAUGGUAAUUAAGUUGACGAUG------------------- ---....((((((((((............))).)).)))))((.....)).((((.((.(((((....))))).....)).))))....------------------- ( -16.80, z-score = 0.48, R) >triCas2.ChLG5 1206362 90 - 18847211 UUUUGCAGUAUUCAGGAUUUCAGCAAACUGCGGAAAAAUGUGCCAUUUGUGGCCAUUUAAUUAUGGAGAUGGUAAACCCUCUUAAAUGGU------------------ (((((((((......(....).....)))))))))(((((.((((....))))))))).......((((.((....)).)))).......------------------ ( -21.70, z-score = -0.78, R) >consensus UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCAUCUGAUCAUGGAAAUGGUUCGUACUCCCCCCA__UCACUC_U__C____C___ .......((((..(((...((.(((((.((((.(....).)))).)))))))...))).(((((....)))))..))))............................. (-15.09 = -14.37 + -0.71)
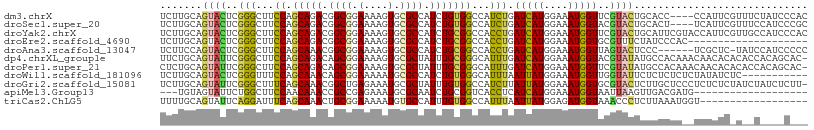
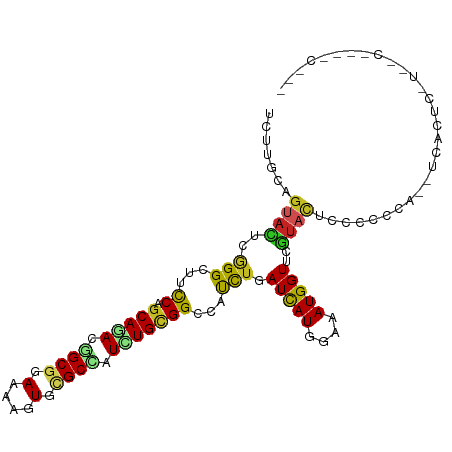
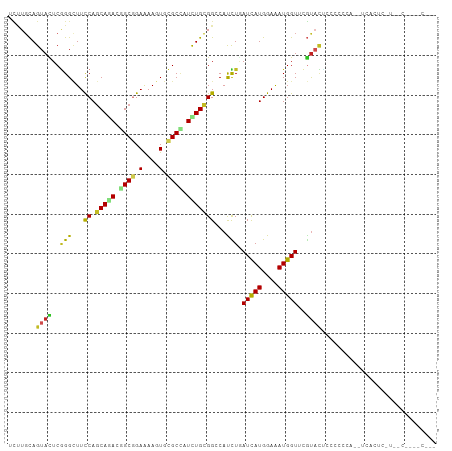
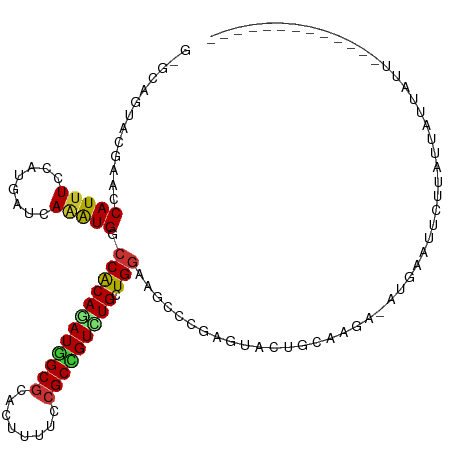
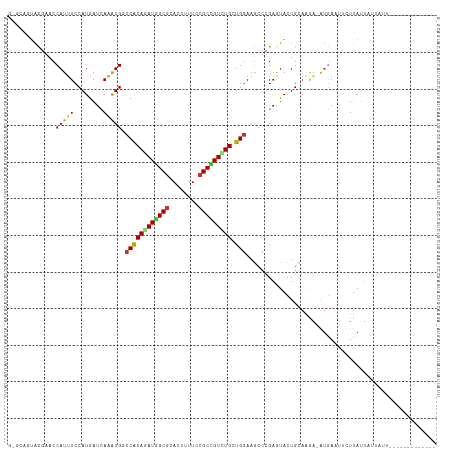
| Location | 13,721,062 – 13,721,178 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.51723 |
| G+C content | 0.44062 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.81 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13721062 116 + 22422827 GUGCAGUACGAACCAUUUCCAUGAUCAGAUGGCCACAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA-GUGAAUUCUUAUUAUUAUUAAUUAAUGGCGUU ..(((((((...((((((........))))))((((((((((((........))))))))).)))........)))))))((((-(....))))).((((((.....)))))).... ( -36.60, z-score = -2.73, R) >droSec1.super_20 368255 116 + 1148123 GUGCAGUACGUACCAUUUCCAUGAUCAGAUGGCCACAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA-GUGAAUUCUUAUUAUUAUUAAUUAAUGGCGUU ..(((((((...((((((........))))))((((((((((((........))))))))).)))........)))))))((((-(....))))).((((((.....)))))).... ( -36.60, z-score = -2.64, R) >droYak2.chrX 8012229 116 + 21770863 AUGCAGUACGAACCAUUUCCAUGAUCAGGUGGCCGCAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA-GGGAAUUCUUAUUAUUAUUAAUUAAUGGCAUU ..(((((((....(((....))).((.(((..((((((((((((........)))))))))).))..))).)))))))))((((-(....))))).((((((.....)))))).... ( -41.40, z-score = -3.27, R) >droEre2.scaffold_4690 5583719 109 - 18748788 -------ACGCACCAUUUCCAUGAUCAGGUGGCCGCAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA-GUGAGUUCUUAUUAUUAUUAAUUAAUGGCGUU -------(((((((((((.((...((.(((..((((((((((((........)))))))))).))..))).))....))...))-))).))......(((((.....))))))))). ( -34.70, z-score = -1.79, R) >dp4.chrXL_group1e 6020984 95 - 12523060 GGCAUAUACGUACCAUUUCCAUGAUCAAAUGCCCGCAAAUAGCGCACUUUUCCGCUGUCUGCUGGAAGCCCGAAUACUGCAGAA-AUGAAUUGUUA--------------------- (((..........(((((........)))))((.(((.((((((........)))))).))).))..)))..((((...((...-.))...)))).--------------------- ( -20.30, z-score = -0.40, R) >droPer1.super_21 134455 95 - 1645244 GGCAUAUACGAACCAUUUCCAUGAUCAAAUGCCCGCAAAUAGCGCACUUUUCCGCUGUCUGCUGGAAGCCCGAAUACUGCAGAG-AUGAAUUGUUA--------------------- .......((((..(((((((((......)))((.(((.((((((........)))))).))).))................)))-)))..))))..--------------------- ( -20.10, z-score = -0.22, R) >droWil1.scaffold_181096 2221036 94 - 12416693 --AGAAUACCAACCAUUUCCAUAAUUAAAUGCCCACAGAUGGCGCAUUUUUCCGCUGUUUGCUGGAAACCCGAGUACUGCAAGA-AUCA--UUUUAUUA------------------ --.............((((((.....(((((((((....))).))))))....((.....))))))))................-....--........------------------ ( -14.40, z-score = -0.01, R) >triCas2.ChLG5 1206372 100 + 18847211 ----AGGGUUUACCAUCUCCAUAAUUAAAUGGCCACAAAUGGCACAUUUUUCCGCAGUUUGCUGAAAUCCUGAAUACUGCAAAACAUCAUUAAUUAUUGGAUUU------------- ----.((.....))...((((((((((((((((((....)))).)))......(((((...(.........)...))))).........))))))).))))...------------- ( -22.10, z-score = -1.66, R) >consensus G_GCAGUACGAACCAUUUCCAUGAUCAAAUGGCCACAGAUGGCGCACUUUUCCGCCGUCUGCUGGAAGCCCGAGUACUGCAAGA_AUGAAUUCUUAUUAUUAUU_____________ .............(((((........))))).((((((((((((........))))))))).))).................................................... (-15.23 = -14.81 + -0.42)
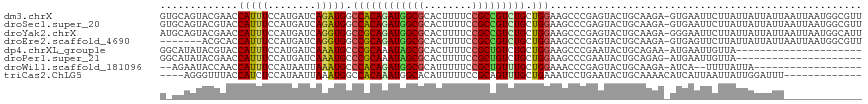
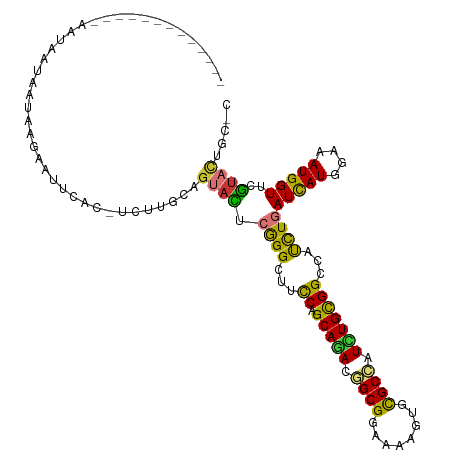
| Location | 13,721,062 – 13,721,178 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.51723 |
| G+C content | 0.44062 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -18.34 |
| Energy contribution | -17.20 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13721062 116 - 22422827 AACGCCAUUAAUUAAUAAUAAUAAGAAUUCAC-UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGUGGCCAUCUGAUCAUGGAAAUGGUUCGUACUGCAC ......................((((......-))))(((((((...((((....(((((.((((........)))).)))))))))....((((((....)))))).))))))).. ( -36.10, z-score = -2.64, R) >droSec1.super_20 368255 116 - 1148123 AACGCCAUUAAUUAAUAAUAAUAAGAAUUCAC-UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGUGGCCAUCUGAUCAUGGAAAUGGUACGUACUGCAC ......................((((......-))))(((((((...((((....(((((.((((........)))).))))))))).....(((((....)))))..))))))).. ( -35.60, z-score = -2.55, R) >droYak2.chrX 8012229 116 - 21770863 AAUGCCAUUAAUUAAUAAUAAUAAGAAUUCCC-UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCACCUGAUCAUGGAAAUGGUUCGUACUGCAU ......................((((......-))))((((((((((((...((.(((((.((((........)))).)))))))...)))))((((....))))...))))))).. ( -38.30, z-score = -2.72, R) >droEre2.scaffold_4690 5583719 109 + 18748788 AACGCCAUUAAUUAAUAAUAAUAAGAACUCAC-UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCACCUGAUCAUGGAAAUGGUGCGU------- ..(((((((............(((((......-)))))((....(((((...((.(((((.((((........)))).)))))))...)))))...))..)))))))...------- ( -33.40, z-score = -1.68, R) >dp4.chrXL_group1e 6020984 95 + 12523060 ---------------------UAACAAUUCAU-UUCUGCAGUAUUCGGGCUUCCAGCAGACAGCGGAAAAGUGCGCUAUUUGCGGGCAUUUGAUCAUGGAAAUGGUACGUAUAUGCC ---------------------...........-..((((((....((.((((((.((.....)))))..))).))....))))))((((.(((((((....))))).))...)))). ( -24.60, z-score = -0.50, R) >droPer1.super_21 134455 95 + 1645244 ---------------------UAACAAUUCAU-CUCUGCAGUAUUCGGGCUUCCAGCAGACAGCGGAAAAGUGCGCUAUUUGCGGGCAUUUGAUCAUGGAAAUGGUUCGUAUAUGCC ---------------------...........-..((((((....((.((((((.((.....)))))..))).))....))))))((((.(((((((....)))).)))...)))). ( -24.60, z-score = -0.30, R) >droWil1.scaffold_181096 2221036 94 + 12416693 ------------------UAAUAAAA--UGAU-UCUUGCAGUACUCGGGUUUCCAGCAAACAGCGGAAAAAUGCGCCAUCUGUGGGCAUUUAAUUAUGGAAAUGGUUGGUAUUCU-- ------------------........--....-......((((((...(((((((((.....))....((((((.(((....))))))))).....)))))))....))))))..-- ( -20.40, z-score = -0.22, R) >triCas2.ChLG5 1206372 100 - 18847211 -------------AAAUCCAAUAAUUAAUGAUGUUUUGCAGUAUUCAGGAUUUCAGCAAACUGCGGAAAAAUGUGCCAUUUGUGGCCAUUUAAUUAUGGAGAUGGUAAACCCU---- -------------...((((.(((((.......(((((((((......(....).....)))))))))(((((.((((....))))))))))))))))))...((....))..---- ( -23.10, z-score = -0.92, R) >consensus _____________AAUAAUAAUAAGAAUUCAC_UCUUGCAGUACUCGGGCUUCCAGCAGACGGCGGAAAAGUGCGCCAUCUGCGGCCAUCUGAUCAUGGAAAUGGUUCGUACUGC_C ........................................((((.((((...((.(((((.((((........)))).)))))))...))))(((((....)))))..))))..... (-18.34 = -17.20 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:23 2011