| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,710,020 – 13,710,123 |
| Length | 103 |
| Max. P | 0.978018 |

| Location | 13,710,020 – 13,710,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.62 |
| Shannon entropy | 0.64833 |
| G+C content | 0.50278 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
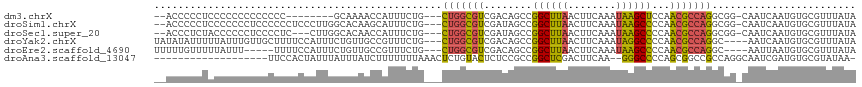
>dm3.chrX 13710020 103 + 22422827 --ACCCCCUCCCCCCCCCCCCC--------GCAAAACCAUUUCUG---CUGGCGUCGACAGCCGGCUUAACUUCAAAUAAGCUCCAACGCCAGGCGG-CAAUCAAUGUGCGUUUAUA --...................(--------(((.....(((.(((---(((((((((.....))(((((........)))))....)))))).))))-.))).....))))...... ( -24.10, z-score = -1.49, R) >droSim1.chrX 10500120 111 + 17042790 --ACCCCCUCCCCCCCUCCCCCCUCCCUUGGCACAAGCAUUUCUG---CUGGCGUCGAUAGCCGGCUUAACUUCAAAUAAGCCCCAACGCCAGGCGG-CAAUCAAUGUGCGUUUAUA --............................(((((...(((.(((---(((((((.(.....)((((((........))))))...)))))).))))-.)))...)))))....... ( -27.60, z-score = -1.43, R) >droSec1.super_20 357397 108 + 1148123 --ACCCUCUACCCCCCUCCCCUC---CUUGGCACAACCAUUUCUG---CUGGCGUCGAUAGCCGGCUUAACUUCAAAUAAGCCCCAACGCCAGGCGG-CAAUCAAUGUGCGUUUAUA --.....................---....(((((...(((.(((---(((((((.(.....)((((((........))))))...)))))).))))-.)))...)))))....... ( -27.60, z-score = -1.80, R) >droYak2.chrX 8001693 110 + 21770863 UAUAUAUUUUUAUUUGUUGCUUUUCCAUUUCUGUUGCCGUUUCUG---CUGGCGUCGACAGCCGGCUUAACUUCAAAUAGGCCCCAACGCCAGGC----AAUCAAUGUGCGUUUAUA ..................((.....((((...(((((((((...(---(((((.......))))))..)))........(((......))).)))----))).)))).))....... ( -27.20, z-score = -1.37, R) >droEre2.scaffold_4690 5572907 105 - 18748788 UUUUUGUUUUUAUUU-----UUUUCCAUUUCUGUUGCCGUUUCUG---CUGGCGUCGACAGCCGGCUUAACUUCAAAUAAGCCCCAACGCCAGGC----AAUUAAUGUGCGUUUAUA ...............-----.....((((...((((((((....)---)((((((.(.....)((((((........))))))...)))))))))----))).)))).......... ( -23.70, z-score = -1.33, R) >droAna3.scaffold_13047 1040699 95 + 1816235 -------------------UUCCACUAUUUAUUUAUCUUUUUUUAAACUCUGUACUCUCCGCCGGCUCGACUUCAA--GGGCCCCAGCGGCCGCCAGGCAAUCGAUGUGCGUAUAA- -------------------................................((((((..(((.(((((........--)))))...)))(((....)))....)).))))......- ( -21.60, z-score = -0.65, R) >consensus __ACCCUCUCCCCCCCUCCCCCCUCCAUUUGCAUAACCAUUUCUG___CUGGCGUCGACAGCCGGCUUAACUUCAAAUAAGCCCCAACGCCAGGCGG_CAAUCAAUGUGCGUUUAUA ................................................(((((((........((((((........))))))...)))))))........................ (-13.86 = -13.50 + -0.36)

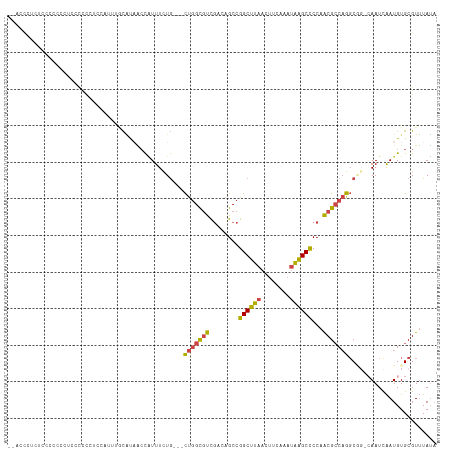
| Location | 13,710,020 – 13,710,123 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.62 |
| Shannon entropy | 0.64833 |
| G+C content | 0.50278 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
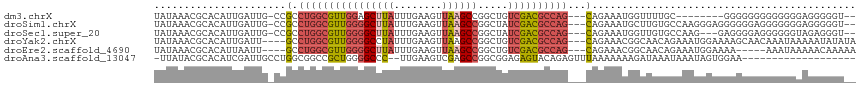
>dm3.chrX 13710020 103 - 22422827 UAUAAACGCACAUUGAUUG-CCGCCUGGCGUUGGAGCUUAUUUGAAGUUAAGCCGGCUGUCGACGCCAG---CAGAAAUGGUUUUGC--------GGGGGGGGGGGGGAGGGGGU-- .....((.(...((..((.-((.(((((((((((((((..((((....))))..)))).)))))))).(---(((((....))))))--------.))).)).))..))..).))-- ( -33.10, z-score = -0.40, R) >droSim1.chrX 10500120 111 - 17042790 UAUAAACGCACAUUGAUUG-CCGCCUGGCGUUGGGGCUUAUUUGAAGUUAAGCCGGCUAUCGACGCCAG---CAGAAAUGCUUGUGCCAAGGGAGGGGGGAGGGGGGGAGGGGGU-- .....((.(.(.....((.-((.(((((((((((((((((........)))))).....)))))))))(---((....)))((.(.((......)).).)))).)).)).)).))-- ( -34.60, z-score = 0.17, R) >droSec1.super_20 357397 108 - 1148123 UAUAAACGCACAUUGAUUG-CCGCCUGGCGUUGGGGCUUAUUUGAAGUUAAGCCGGCUAUCGACGCCAG---CAGAAAUGGUUGUGCCAAG---GAGGGGAGGGGGGUAGAGGGU-- ................(((-((.(((((((((((((((((........)))))).....))))))))..---......(((.....)))..---.......))).))))).....-- ( -32.80, z-score = 0.08, R) >droYak2.chrX 8001693 110 - 21770863 UAUAAACGCACAUUGAUU----GCCUGGCGUUGGGGCCUAUUUGAAGUUAAGCCGGCUGUCGACGCCAG---CAGAAACGGCAACAGAAAUGGAAAAGCAACAAAUAAAAAUAUAUA .......((.......((----((.(((((((((((((..((((....))))..)))).))))))))))---))).........((....)).....)).................. ( -27.60, z-score = -1.05, R) >droEre2.scaffold_4690 5572907 105 + 18748788 UAUAAACGCACAUUAAUU----GCCUGGCGUUGGGGCUUAUUUGAAGUUAAGCCGGCUGUCGACGCCAG---CAGAAACGGCAACAGAAAUGGAAAA-----AAAUAAAAACAAAAA ..........((((..((----((.(((((((((((((((........)))))).....))))))))))---)))....(....)...)))).....-----............... ( -27.70, z-score = -2.00, R) >droAna3.scaffold_13047 1040699 95 - 1816235 -UUAUACGCACAUCGAUUGCCUGGCGGCCGCUGGGGCCC--UUGAAGUCGAGCCGGCGGAGAGUACAGAGUUUAAAAAAAGAUAAAUAAAUAGUGGAA------------------- -.....(((((.((..(((((.(((((((.....)))).--(((....))))))))))).)))).....(((((........))))).....)))...------------------- ( -22.20, z-score = 0.58, R) >consensus UAUAAACGCACAUUGAUUG_CCGCCUGGCGUUGGGGCUUAUUUGAAGUUAAGCCGGCUGUCGACGCCAG___CAGAAAUGGUUAUACAAAUGGAGAAGGGAGGGGGGAAGAGGGU__ ........................((((((((((((((..((((....))))..)))).))))))))))................................................ (-16.41 = -16.75 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:19 2011