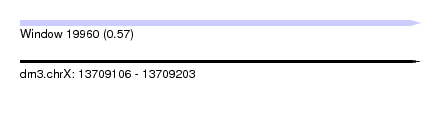
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,709,106 – 13,709,203 |
| Length | 97 |
| Max. P | 0.568378 |

| Location | 13,709,106 – 13,709,203 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.64 |
| Shannon entropy | 0.47710 |
| G+C content | 0.41755 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -9.65 |
| Energy contribution | -11.81 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
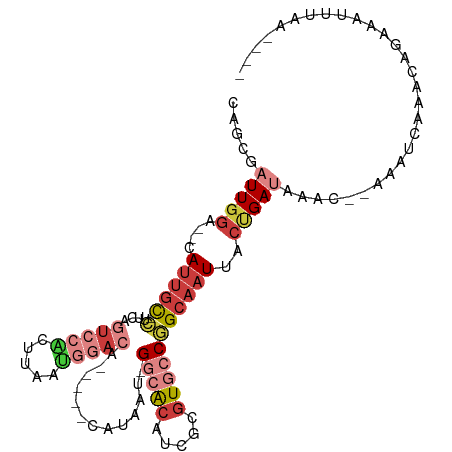
>dm3.chrX 13709106 97 + 22422827 CAGCGAUUGGA--CAUUGC-CUCCAGUCCACUUAAUGGACA-----CAUAAU-GGCACAUCGCGUGCCGGCAAUUACUGAUAAACGCAAAUCAAACAGAAAUUUAA---- ..(((((..(.--.(((((-(....(((((.....))))).-----......-(((((.....)))))))))))..)..))...)))...................---- ( -28.60, z-score = -3.67, R) >droSim1.chrX 10499273 97 + 17042790 CAGCGAUUGGA--CAUUGC-CUUCAGUCCACUUAAUGAACA-----CAUAAU-GGCACAUCGCGUGCCGGCAAUUACUGAUAAACACAAAUCAAACAGAAAUUUAA---- .....((..(.--.(((((-(((((..........))))..-----......-(((((.....)))))))))))..)..)).........................---- ( -18.90, z-score = -1.23, R) >droSec1.super_20 356534 97 + 1148123 CAGCGAUUGGA--CAUUGC-CUUCAGUCCACUUAAUGGACA-----CAUAAU-GGCACAUCGCGUGCCGGCAAUUACUGAUAAACACAAAUCAAACAGAAAUUUAA---- .....((..(.--.(((((-(....(((((.....))))).-----......-(((((.....)))))))))))..)..)).........................---- ( -25.20, z-score = -3.15, R) >droYak2.chrX 8000700 95 + 21770863 CAGCGAUUGGA--CAUUGC-CUCCUGUCCACUUAAUGGACA-----CAUAAU-GGCACAUCGCGUGCCGGCAAUUACUGAUAAAU--AAAUCGAACACAAAUUUAA---- .....((..(.--.(((((-(...((((((.....))))))-----......-(((((.....)))))))))))..)..))....--...................---- ( -25.50, z-score = -3.01, R) >droEre2.scaffold_4690 5571904 95 - 18748788 CAGCGAUUGGA--GAUUGU-CUUCUGUCCGAUUAAUGAACA-----CAUAAU-GGCACAUCGCGUGCCGGCAAUUACUGAUAAAU--AAAUCAAACAGAAAUUUAA---- ..(((((((((--.(....-....).)))))))........-----.....(-(((((.....))))))))......((((....--..)))).............---- ( -18.80, z-score = -0.62, R) >droAna3.scaffold_13047 1040130 99 + 1816235 CAACGCUUGGA--CAGCGUGAACCACUCCCCCUCGGAGCCAGCUUGCAUAAU-GAGGCUUCGCGUGCCGGCAAUUACCGAUAAAC--AAAUCAAACAAAUUCAA------ ....(((.((.--..((((((....((((.....))))..(((((.......-.))))))))))).))))).......(((....--..)))............------ ( -22.10, z-score = -0.28, R) >droWil1.scaffold_181096 9357638 108 + 12416693 CAGUUUUUCAUUUCAUUUCAUUUGAUUCCCCCUCCCAGAUCCCUCCCACCUUUGAAAAUUCGCGUGCCAGCAAUUACUGAUAAAU--AAAUCAAACAAAAAAGCUCUUAA ((((((((((.........(((((...........)))))............))))))...((......))....))))......--....................... ( -6.90, z-score = 0.56, R) >consensus CAGCGAUUGGA__CAUUGC_CUUCAGUCCACUUAAUGGACA_____CAUAAU_GGCACAUCGCGUGCCGGCAAUUACUGAUAAAC__AAAUCAAACAGAAAUUUAA____ (((..((((................(((((.....))))).............(((((.....)))))..))))..)))............................... ( -9.65 = -11.81 + 2.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:16 2011