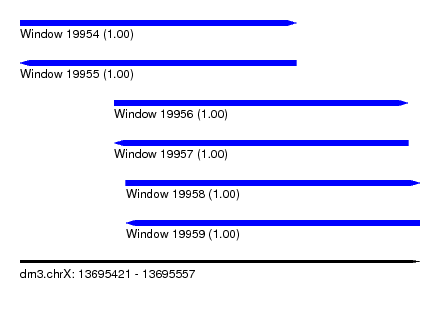
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,695,421 – 13,695,557 |
| Length | 136 |
| Max. P | 0.999996 |

| Location | 13,695,421 – 13,695,515 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.04 |
| Shannon entropy | 0.61864 |
| G+C content | 0.38959 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
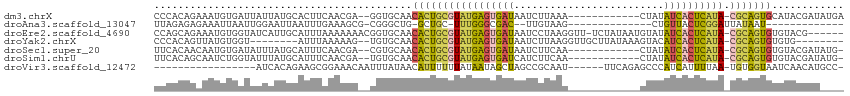
>dm3.chrX 13695421 94 + 22422827 AUCAUGAUUGCUUUU----GGUGUCAGGAUCUUUCCCGGAUCAUAUCGUAUGCACUGCGUAUGAGUGAUAUAGUUUA------------AGAUUAUCACUCAUACGCAGU .........((...(----(((((...(((((.....)))))))))))...))(((((((((((((((((.......------------....))))))))))))))))) ( -36.10, z-score = -5.15, R) >droEre2.scaffold_4690 5558127 98 - 18748788 AUCAUAAUUGUUUUC----GUUGCCAGGAUCAGGCACAG-------CGUACACACUGCGUAUGAGUGAUAUACAUUAUAGA-AACCUUAGGAUUAUCACUCAUACGCAGU ........(((...(----((((((.......))))..)-------)).))).(((((((((((((((((....(((.((.-...)))))...))))))))))))))))) ( -35.00, z-score = -5.69, R) >droYak2.chrX 7987074 101 + 21770863 AUCAUGAUUGUUUUUUUUAGUUGCCAGGAUCUGUAAAGG-------C--ACACACUGCGUAUGAGUGAUGUACUUUAUAAGCAACCUUAAGAUUAUCACUCAUACGCAGU ...................((((((............))-------)--).))(((((((((((((((((....((.((((....)))).)).))))))))))))))))) ( -32.50, z-score = -4.31, R) >droSec1.super_20 342935 92 + 1148123 AUCAUAAUUGCUUUU----GGUGCCACCAUCUUUCCAGG--CAUAUCGUACACACUGCGUAUGAGUGAUAUAGUUGA------------AGAUUAUCACUCAUACGCAGU ........(((....----.(((((............))--)))...)))...(((((((((((((((((.......------------....))))))))))))))))) ( -32.60, z-score = -4.98, R) >droSim1.chrU 1389434 92 - 15797150 AUCAUGAUUGCUUUU----GGUGCCACGAUCUUUCCAGG--CAUAUCGUACACACUGCGUAUGAGUGAUAUAGUUGA------------AGAUGAUCACUCAUACGCAGU ...(((((((((..(----((.............)))))--)).)))))....((((((((((((((((...(((..------------.))).)))))))))))))))) ( -33.42, z-score = -3.82, R) >droVir3.scaffold_12472 239053 99 + 763072 AUUCUGAUUAAUGUUUC--GUUCCGAGAAUUGUCCAUCG---GCAUGUUGAUUACCACAUUAAAAUGAUGGGCUCUGAAA------UUGCGGCUAGCUAUUAUAAAAAAU .(((.((......((((--(...)))))...((((((((---.....(((((......)))))..)))))))))).))).------..((.....))............. ( -15.80, z-score = 0.84, R) >consensus AUCAUGAUUGCUUUU____GGUGCCAGGAUCUUUCAAGG___AUAUCGUACACACUGCGUAUGAGUGAUAUACUUUA____________AGAUUAUCACUCAUACGCAGU .....................................................(((((((((((((((((.......................))))))))))))))))) (-19.04 = -20.43 + 1.39)

| Location | 13,695,421 – 13,695,515 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.04 |
| Shannon entropy | 0.61864 |
| G+C content | 0.38959 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13695421 94 - 22422827 ACUGCGUAUGAGUGAUAAUCU------------UAAACUAUAUCACUCAUACGCAGUGCAUACGAUAUGAUCCGGGAAAGAUCCUGACACC----AAAAGCAAUCAUGAU (((((((((((((((((....------------.......)))))))))))))))))((....(((...)))(((((....))))).....----....))......... ( -32.50, z-score = -5.65, R) >droEre2.scaffold_4690 5558127 98 + 18748788 ACUGCGUAUGAGUGAUAAUCCUAAGGUU-UCUAUAAUGUAUAUCACUCAUACGCAGUGUGUACG-------CUGUGCCUGAUCCUGGCAAC----GAAAACAAUUAUGAU (((((((((((((((((...........-...........)))))))))))))))))((...((-------...((((.......)))).)----)...))......... ( -32.95, z-score = -4.40, R) >droYak2.chrX 7987074 101 - 21770863 ACUGCGUAUGAGUGAUAAUCUUAAGGUUGCUUAUAAAGUACAUCACUCAUACGCAGUGUGU--G-------CCUUUACAGAUCCUGGCAACUAAAAAAAACAAUCAUGAU ((((((((((((((((...((((((....))))...))...))))))))))))))))...(--(-------((............))))..................... ( -32.00, z-score = -4.61, R) >droSec1.super_20 342935 92 - 1148123 ACUGCGUAUGAGUGAUAAUCU------------UCAACUAUAUCACUCAUACGCAGUGUGUACGAUAUG--CCUGGAAAGAUGGUGGCACC----AAAAGCAAUUAUGAU (((((((((((((((((....------------.......)))))))))))))))))..........((--(((....)).(((.....))----)...)))........ ( -32.00, z-score = -4.23, R) >droSim1.chrU 1389434 92 + 15797150 ACUGCGUAUGAGUGAUCAUCU------------UCAACUAUAUCACUCAUACGCAGUGUGUACGAUAUG--CCUGGAAAGAUCGUGGCACC----AAAAGCAAUCAUGAU ((((((((((((((((.....------------........))))))))))))))))(((((((((...--.((....))))))).)))).----............... ( -32.82, z-score = -4.12, R) >droVir3.scaffold_12472 239053 99 - 763072 AUUUUUUAUAAUAGCUAGCCGCAA------UUUCAGAGCCCAUCAUUUUAAUGUGGUAAUCAACAUGC---CGAUGGACAAUUCUCGGAAC--GAAACAUUAAUCAGAAU .............((.....))..------((((.((..(((.(((....))))))...))......(---(((.(((...)))))))...--))))............. ( -11.40, z-score = 1.38, R) >consensus ACUGCGUAUGAGUGAUAAUCU____________UAAACUAUAUCACUCAUACGCAGUGUGUACGAUAU___CCUGGAAAGAUCCUGGCAAC____AAAAGCAAUCAUGAU ((((((((((((((((.........................))))))))))))))))..................................................... (-19.05 = -19.53 + 0.48)
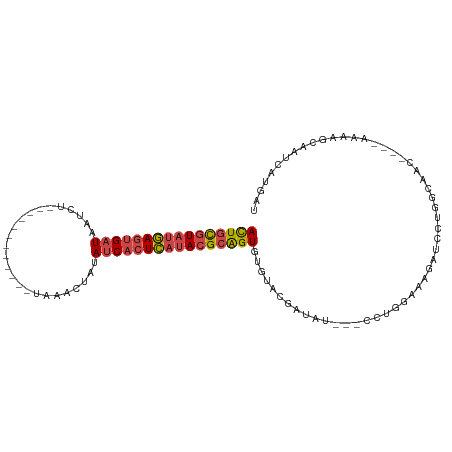
| Location | 13,695,453 – 13,695,553 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 48.35 |
| Shannon entropy | 0.89336 |
| G+C content | 0.38974 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -11.02 |
| Energy contribution | -14.88 |
| Covariance contribution | 3.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.37 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13695453 100 + 22422827 CGGAUCAUAUCGUAUGCACUGCGUAUGAGUGAUAUAG-----------UUUAAGAUUAUCACUCAUACGCAGU-GUUGCACC--UCGUUGAAGUGCAUAAUAAUCACAUUUCUG----- ((((...........(((((((((((((((((((...-----------........)))))))))))))))))-))(((((.--((...)).))))).............))))----- ( -38.70, z-score = -5.54, R) >droAna3.scaffold_13047 1025854 82 + 1816235 ---------------------------GGGAUUAUAAUCCG-----AGUAACAGCUUACAAGUCGCCCAAAGCAGCCAGCCCGCGCUUUCAAAUUAAUUCCAAUUAAUUUCUCU----- ---------------------------(((((((......(-----(((....))))...........(((((.((......))))))).....))))))).............----- ( -12.80, z-score = -0.70, R) >droEre2.scaffold_4690 5558159 106 - 18748788 -------CAGCGUACACACUGCGUAUGAGUGAUAUACAUUAUAGAAACCUUAGGAUUAUCACUCAUACGCAGU-GUUGCACCGUUUUUUUAAAUGCAAUGAUACCACAUUUCUG----- -------..(((((.(((((((((((((((((((....(((.((....)))))...)))))))))))))))))-)))))...((((....))))))..................----- ( -35.40, z-score = -5.90, R) >droSec1.super_20 342967 98 + 1148123 --AGGCAUAUCGUACACACUGCGUAUGAGUGAUAUAG-----------UUGAAGAUUAUCACUCAUACGCAGU-GUUGCACG--UCGUUGAAAUGCAUAAAUAUCACAUUGUUG----- --..((((...(((.(((((((((((((((((((...-----------........)))))))))))))))))-))))).((--....))..))))..................----- ( -36.60, z-score = -4.26, R) >droSim1.chrU 1389466 98 - 15797150 --AGGCAUAUCGUACACACUGCGUAUGAGUGAUAUAG-----------UUGAAGAUGAUCACUCAUACGCAGU-GUUGCACA--UCGUUGAAAUGCAUAAAUACCAGAUUGCUG----- --.((((.((((((.((((((((((((((((((...(-----------((...))).))))))))))))))))-)))))...--..((......))..........))))))).----- ( -38.60, z-score = -4.97, R) >droVir3.scaffold_12472 239086 89 + 763072 ------------------------AUCGGCAUGUUGAUUACC----ACAUUAAAAUGAUGGGC--UCUGAAAUUGCGGCUAGCUAUUAUAAAAAAUGUUAUAAAUUGUUUCCGCUUCUG ------------------------.((((...(((.((((..----.........)))).)))--.))))....((((..(((..((((((......))))))...))).))))..... ( -14.20, z-score = 0.21, R) >consensus _______UAUCGUACACACUGCGUAUGAGUGAUAUAG___________UUUAAGAUUAUCACUCAUACGCAGU_GUUGCACC__UCGUUGAAAUGCAUAAAAAUCACAUUCCUG_____ .................(((((((((((((((((......................))))))))))))))))).............................................. (-11.02 = -14.88 + 3.86)

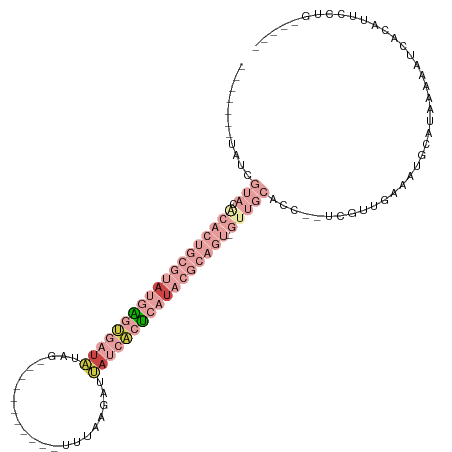
| Location | 13,695,453 – 13,695,553 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 48.35 |
| Shannon entropy | 0.89336 |
| G+C content | 0.38974 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -10.16 |
| Energy contribution | -15.17 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.35 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.71 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13695453 100 - 22422827 -----CAGAAAUGUGAUUAUUAUGCACUUCAACGA--GGUGCAAC-ACUGCGUAUGAGUGAUAAUCUUAAA-----------CUAUAUCACUCAUACGCAGUGCAUACGAUAUGAUCCG -----.......(.((((((..((((((((...))--)))))).(-(((((((((((((((((........-----------...))))))))))))))))))........))))))). ( -42.30, z-score = -7.57, R) >droAna3.scaffold_13047 1025854 82 - 1816235 -----AGAGAAAUUAAUUGGAAUUAAUUUGAAAGCGCGGGCUGGCUGCUUUGGGCGACUUGUAAGCUGUUACU-----CGGAUUAUAAUCCC--------------------------- -----.(((((((((((....))))))))...((((((((.(.(((......))).))))))..)))....))-----)((((....)))).--------------------------- ( -17.40, z-score = 0.14, R) >droEre2.scaffold_4690 5558159 106 + 18748788 -----CAGAAAUGUGGUAUCAUUGCAUUUAAAAAAACGGUGCAAC-ACUGCGUAUGAGUGAUAAUCCUAAGGUUUCUAUAAUGUAUAUCACUCAUACGCAGUGUGUACGCUG------- -----((((((((..((...))..))))).........(((((.(-(((((((((((((((((......................))))))))))))))))))))))).)))------- ( -41.05, z-score = -6.33, R) >droSec1.super_20 342967 98 - 1148123 -----CAACAAUGUGAUAUUUAUGCAUUUCAACGA--CGUGCAAC-ACUGCGUAUGAGUGAUAAUCUUCAA-----------CUAUAUCACUCAUACGCAGUGUGUACGAUAUGCCU-- -----....(((((.........))))).......--((((((.(-(((((((((((((((((........-----------...))))))))))))))))))))))))........-- ( -39.30, z-score = -6.32, R) >droSim1.chrU 1389466 98 + 15797150 -----CAGCAAUCUGGUAUUUAUGCAUUUCAACGA--UGUGCAAC-ACUGCGUAUGAGUGAUCAUCUUCAA-----------CUAUAUCACUCAUACGCAGUGUGUACGAUAUGCCU-- -----..((((((..(((....)))..........--.(((((.(-((((((((((((((((.........-----------....))))))))))))))))))))))))).)))..-- ( -38.32, z-score = -5.48, R) >droVir3.scaffold_12472 239086 89 - 763072 CAGAAGCGGAAACAAUUUAUAACAUUUUUUAUAAUAGCUAGCCGCAAUUUCAGA--GCCCAUCAUUUUAAUGU----GGUAAUCAACAUGCCGAU------------------------ .....((((.......((((((......)))))).......)))).......((--..(((.(((....))))----))...))...........------------------------ ( -13.64, z-score = -0.53, R) >consensus _____CAGAAAUGUGAUAUUAAUGCAUUUCAAAGA__GGUGCAAC_ACUGCGUAUGAGUGAUAAUCUUAAA___________CUAUAUCACUCAUACGCAGUGUGUACGAUA_______ ......................(((((...........)))))...(((((((((((((((((......................)))))))))))))))))................. (-10.16 = -15.17 + 5.00)

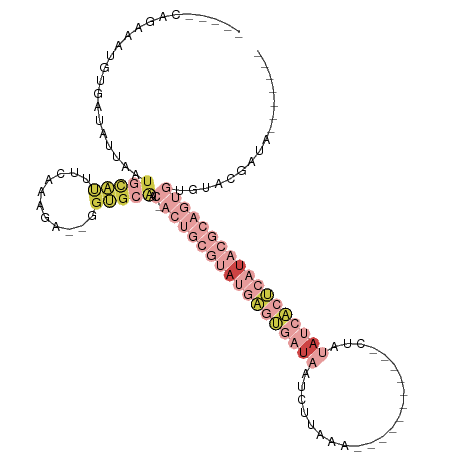
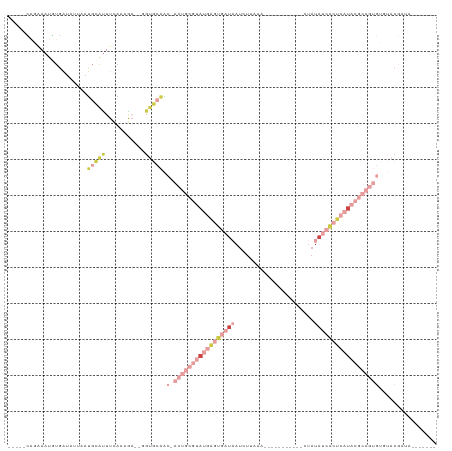
| Location | 13,695,457 – 13,695,557 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 52.09 |
| Shannon entropy | 0.93005 |
| G+C content | 0.38119 |
| Mean single sequence MFE | -30.21 |
| Consensus MFE | -14.69 |
| Energy contribution | -16.63 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 6.24 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13695457 100 + 22422827 UCAUAUCGUAUGCACUGCG-UAUGAGUGAUAUAG------------UUUAAGAUUAUCACUCAUACGCAGUGUUGCACC--UCGUUGAAGUGCAUAAUAAUCACAUUUCUGUGGG .......(((.((((((((-(((((((((((...------------........))))))))))))))))))))))..(--(((..((((((...........))))))..)))) ( -40.50, z-score = -5.93, R) >droAna3.scaffold_13047 1025857 83 + 1816235 -------------AUUAUAAUCCGAGUAACAG--------------CUUACAA--GUCGCCCAAA-GCAGC-CAGCCCG-CGCUUUCAAAUUAAUUCCAAUUAAUUUCUCUCUAA -------------..........(((......--------------.......--.......(((-((.((-......)-)))))).((((((((....)))))))))))..... ( -8.50, z-score = -0.65, R) >droEre2.scaffold_4690 5558162 107 - 18748788 ------CGUACACACUGCG-UAUGAGUGAUAUACAUUAUAGA-AACCUUAGGAUUAUCACUCAUACGCAGUGUUGCACCGUUUUUUUAAAUGCAAUGAUACCACAUUUCUGCUGG ------.(((.((((((((-(((((((((((....(((.((.-...)))))...))))))))))))))))))))))...............(((..(((.....)))..)))... ( -36.10, z-score = -5.60, R) >droYak2.chrX 7987113 96 + 21770863 --------CACACACUGCG-UAUGAGUGAUGUACUUUAUAAGCAACCUUAAGAUUAUCACUCAUACGCAGUGUUGCACA--CUUUUUAAAU--------ACCACAUAACUGUGGG --------...((((((((-(((((((((((....((.((((....)))).)).)))))))))))))))))))......--..........--------.(((((....))))). ( -38.30, z-score = -7.53, R) >droSec1.super_20 342970 99 + 1148123 -CAUAUCGUACACACUGCG-UAUGAGUGAUAUAG------------UUGAAGAUUAUCACUCAUACGCAGUGUUGCACG--UCGUUGAAAUGCAUAAAUAUCACAUUGUUGUGAA -..........((((((((-(((((((((((...------------........)))))))))))))))))))((((..--((...))..))))......(((((....))))). ( -39.40, z-score = -4.99, R) >droSim1.chrU 1389469 99 - 15797150 -CAUAUCGUACACACUGCG-UAUGAGUGAUAUAG------------UUGAAGAUGAUCACUCAUACGCAGUGUUGCACA--UCGUUGAAAUGCAUAAAUACCAGAUUGCUGUGAA -((((..(((.((((((((-((((((((((...(------------((...))).)))))))))))))))))))))...--..........(((............))))))).. ( -35.00, z-score = -3.70, R) >droVir3.scaffold_12472 239089 90 + 763072 -GGCAUGUUGAUUACCACA-UUAAAAUGAUGGGCUCUGAA------AUUGCGGCUAGCUAUUAUAAAAAAUGUUAUAAAUUGUUUCCGCUUCUGUGAU----------------- -.(((.........(((((-(....))).)))........------...((((..(((..((((((......))))))...))).))))...)))...----------------- ( -13.70, z-score = 0.69, R) >consensus ____AUCGUACACACUGCG_UAUGAGUGAUAUAC____________CUUAAGAUUAUCACUCAUACGCAGUGUUGCACA__UCUUUGAAAUGCAUAAAUACCACAUUGCUGUGAA ...........((((((((.(((((((((((.......................))))))))))))))))))).......................................... (-14.69 = -16.63 + 1.94)
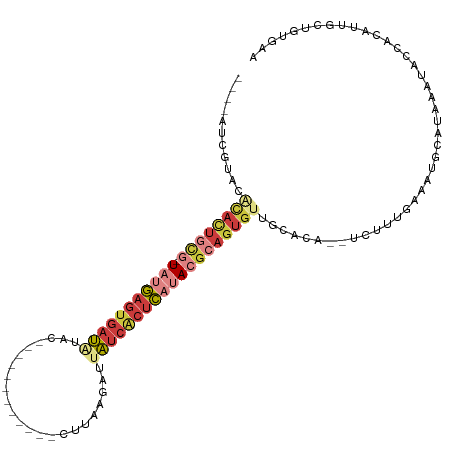
| Location | 13,695,457 – 13,695,557 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 52.09 |
| Shannon entropy | 0.93005 |
| G+C content | 0.38119 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.65 |
| Mean z-score | -4.89 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 6.46 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
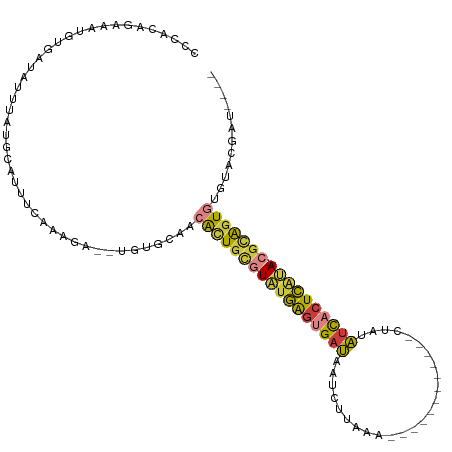
>dm3.chrX 13695457 100 - 22422827 CCCACAGAAAUGUGAUUAUUAUGCACUUCAACGA--GGUGCAACACUGCGUAUGAGUGAUAAUCUUAAA------------CUAUAUCACUCAUA-CGCAGUGCAUACGAUAUGA ..((((....)))).......((((((((...))--)))))).((((((((((((((((((........------------...)))))))))))-)))))))............ ( -44.30, z-score = -8.55, R) >droAna3.scaffold_13047 1025857 83 - 1816235 UUAGAGAGAAAUUAAUUGGAAUUAAUUUGAAAGCG-CGGGCUG-GCUGC-UUUGGGCGAC--UUGUAAG--------------CUGUUACUCGGAUUAUAAU------------- ........((((((((....)))))))).......-((((.((-((.((-((.((....)--)...)))--------------).)))))))).........------------- ( -16.00, z-score = 0.08, R) >droEre2.scaffold_4690 5558162 107 + 18748788 CCAGCAGAAAUGUGGUAUCAUUGCAUUUAAAAAAACGGUGCAACACUGCGUAUGAGUGAUAAUCCUAAGGUU-UCUAUAAUGUAUAUCACUCAUA-CGCAGUGUGUACG------ .......(((((..((...))..))))).........(((((.((((((((((((((((((...........-...........)))))))))))-)))))))))))).------ ( -40.15, z-score = -6.03, R) >droYak2.chrX 7987113 96 - 21770863 CCCACAGUUAUGUGGU--------AUUUAAAAAG--UGUGCAACACUGCGUAUGAGUGAUAAUCUUAAGGUUGCUUAUAAAGUACAUCACUCAUA-CGCAGUGUGUG-------- .(((((....))))).--------..........--...(((.(((((((((((((((((...((((((....))))...))...))))))))))-)))))))))).-------- ( -38.80, z-score = -5.94, R) >droSec1.super_20 342970 99 - 1148123 UUCACAACAAUGUGAUAUUUAUGCAUUUCAACGA--CGUGCAACACUGCGUAUGAGUGAUAAUCUUCAA------------CUAUAUCACUCAUA-CGCAGUGUGUACGAUAUG- .(((((....)))))...................--((((((.((((((((((((((((((........------------...)))))))))))-))))))))))))).....- ( -42.30, z-score = -7.35, R) >droSim1.chrU 1389469 99 + 15797150 UUCACAGCAAUCUGGUAUUUAUGCAUUUCAACGA--UGUGCAACACUGCGUAUGAGUGAUCAUCUUCAA------------CUAUAUCACUCAUA-CGCAGUGUGUACGAUAUG- .((.(((....)))((((...(((((.((...))--.)))))((((((((((((((((((.........------------....))))))))))-))))))))))))))....- ( -36.62, z-score = -5.08, R) >droVir3.scaffold_12472 239089 90 - 763072 -----------------AUCACAGAAGCGGAAACAAUUUAUAACAUUUUUUAUAAUAGCUAGCCGCAAU------UUCAGAGCCCAUCAUUUUAA-UGUGGUAAUCAACAUGCC- -----------------((((((...((((.......((((((......)))))).......))))...------....((.....)).......-))))))............- ( -14.84, z-score = -1.34, R) >consensus CCCACAGAAAUGUGAUAUUUAUGCAUUUCAAAGA__UGUGCAACACUGCGUAUGAGUGAUAAUCUUAAA____________CUAUAUCACUCAUA_CGCAGUGUGUACGAU____ ...........................................(((((((((((((((((.........................)))))))))).)))))))............ (-15.26 = -15.42 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:15 2011