
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,681,677 – 13,681,813 |
| Length | 136 |
| Max. P | 0.584954 |

| Location | 13,681,677 – 13,681,773 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.51745 |
| G+C content | 0.51763 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -9.02 |
| Energy contribution | -9.40 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
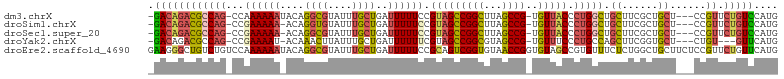
>dm3.chrX 13681677 96 - 22422827 ---------GCUAAGCCGGCUACGGAAAAAUCAGCAAAUACGCCUGUAUUUUUUGG-CUGGCGUCUGUCGCACUCCCUGCCCUUUCCCAUUUCCCUUUUGCAUCCU-- ---------((...(((((((...(((((((((((......).))).)))))))))-))))).......(((.....)))...................)).....-- ( -20.80, z-score = -0.49, R) >droSim1.chrX 10468567 96 - 17042790 ---------GCUAAGCCGGCUACGGAAAAAUCAGCAAAUACACCUGU-UUUUUCGG-CUGGCGUCUGUCGCACU-CCUGCCCUUUCCCAUUUCCCUUUGGCAUCCCCU ---------(((((((((((..((((((((.(((.........))))-))))))))-))))).......(((..-..)))................)))))....... ( -24.60, z-score = -1.62, R) >droYak2.chrX 7973727 92 - 21770863 ---------GCUACGCCGGCUACGAAAAAAUCAGCAAAUAAGUUUGU-AUUUUCGG-CUGGCGUCUGUCGUACUCC---CCCCUCUCCACCCCUUCGUCACAAAUU-- ---------((.((((((((..((((((.((.(((......))).))-.)))))))-)))))))..))........---...........................-- ( -20.10, z-score = -2.30, R) >droEre2.scaffold_4690 5545629 104 + 18748788 GCUACACCGGUUACACCGACUGCGGAAAAAUCAGCAAAUACGCCUGUAUUUUUUGGACAGACAGCCCUUCAGCAUCC--CCACCAACCACUCCACUCACCCCACCU-- (((....(((.....))).((((.(((((((((((......).))).))))))).).)))..........)))....--...........................-- ( -17.40, z-score = -1.57, R) >consensus _________GCUAAGCCGGCUACGGAAAAAUCAGCAAAUACGCCUGU_UUUUUCGG_CUGGCGUCUGUCGCACUCCC__CCCCUUCCCACUCCCCUUUCGCAACCU__ ..............(((((...((((((...(((.........)))...))))))..))))).............................................. ( -9.02 = -9.40 + 0.38)



| Location | 13,681,714 – 13,681,813 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Shannon entropy | 0.31062 |
| G+C content | 0.52554 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.76 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13681714 99 + 22422827 -GACAGACGCCAG-CCAAAAAAUACAGGCGUAUUUGCUGAUUUUUCCGUAGCCGGCUUAGCCG-UGUUACCCUGGCUGCUUCGCUGCU---CCGUUCUGUCCAUG -((((((((.(((-(..((((((...((((....)))).))))))..((((((((..((((..-.))))..))))))))...))))..---.)).)))))).... ( -33.70, z-score = -2.94, R) >droSim1.chrX 10468605 98 + 17042790 -GACAGACGCCAG-CCGAAAAA-ACAGGUGUAUUUGCUGAUUUUUCCGUAGCCGGCUUAGCCG-UGUUACCCUGGCUGCUUCGCUGCU---CCGUUCUGUCCAUG -((((((((.(((-(.((((((-.(((.........))).)))))).((((((((..((((..-.))))..))))))))...))))..---.)).)))))).... ( -33.80, z-score = -2.98, R) >droSec1.super_20 320259 98 + 1148123 -GACAGACGCCAG-CCGAAAAA-ACAGGCGUAUUUGCUGAUUUUUCCGUAGCCGGCUUAGCCG-UGUUACCCUGGCUGCUUCGCUGCU---CCGUUCUGUCCAUG -((((((((.(((-(.((((((-...((((....))))..)))))).((((((((..((((..-.))))..))))))))...))))..---.)).)))))).... ( -35.50, z-score = -3.25, R) >droYak2.chrX 7973761 95 + 21770863 -GACAGACGCCAG-CCGAAAAU-ACAAACUUAUUUGCUGAUUUUUUCGUAGCCGGCGUAGCCG-UGUUUCCCUGCCAGCUUCGGUGCU---CUGU---GUUCAUG -.(((((((((.(-(((.((((-(......)))))((((.........))))))))(.(((.(-.((......))).))).))))).)---))))---....... ( -24.30, z-score = -0.74, R) >droEre2.scaffold_4690 5545663 105 - 18748788 GAAGGGCUGUCUGUCCAAAAAAUACAGGCGUAUUUGCUGAUUUUUCCGCAGUCGGUGUAACCGGUGUAGCCGUGUUUCUCUGGCUGCUUCUCCGUUCUGUUCAUG (((((((.....)))).((((((...((((....)))).))))))..((((((((.(.((((((.....))).))).).))))))))............)))... ( -31.90, z-score = -2.44, R) >consensus _GACAGACGCCAG_CCGAAAAA_ACAGGCGUAUUUGCUGAUUUUUCCGUAGCCGGCUUAGCCG_UGUUACCCUGGCUGCUUCGCUGCU___CCGUUCUGUCCAUG .((((((((((((...((((((....((((....))))..)))))).(((((((((...))))..))))).))))).((......))......)).))))).... (-14.68 = -15.76 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:08 2011