| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,652,909 – 13,653,008 |
| Length | 99 |
| Max. P | 0.729458 |

| Location | 13,652,909 – 13,653,008 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.81 |
| Shannon entropy | 0.60864 |
| G+C content | 0.45609 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729458 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
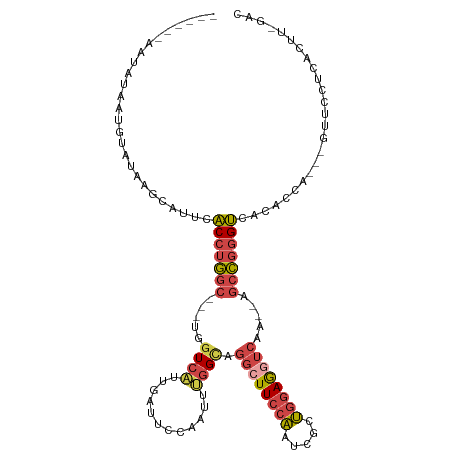
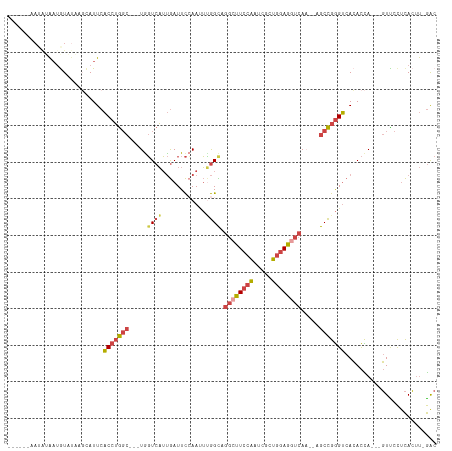
>dm3.chrX 13652909 99 + 22422827 -----------AAUAUA-AUGCAUUCACCUGGCGGCUGGUCAUUGAUUCCAAUUUGGCCGGCUUCCAAUCGCUGGAGGUCAA--UGCCGGGUCACACCA---GUUCCUCACUU-GAC -----------......-........((((((((((..(..((((....)))))..)))((((((((.....))))))))..--.))))))).......---...........-... ( -32.50, z-score = -1.75, R) >droSim1.chrX_random 3723451 110 + 5698898 ------AAUAUAAUGUAUAAGCAUUCACCUGGCGGCUGGUCAUUGAUUCCAAUUUGGCAGGCUUCCAAUCGCUGGAGGUCACACAGCCGGGUCACACCACCAGUUCCUCACUU-GAC ------.....(((((....)))))...(((((((((((((((((....)))..)))).((((((((.....))))))))...))))))((.....)).))))..........-... ( -33.50, z-score = -1.73, R) >droSec1.super_20 291631 111 + 1148123 ------AAUGCAAUGUAUAAGCAUUCACCUGGCGGCUGGUCAUUGAUUCCAAUUUGGCAGGCUUCCAAUCGCUGGAGGUCACACUGCCGGGUCACACCACCAGUUCCUCACUUUGAC ------(((((.........))))).(((((((((.(((((((((....)))..)))).((((((((.....)))))))).)))))))))))......................... ( -33.20, z-score = -0.93, R) >droYak2.chrX 7945854 106 + 21770863 --UGUAAAUAUACUGUAUACUUAUUCACCUGGC---UGGUCAUUGAUUCCAACUCGGUCGGCUUCCAAUCGCUGGAGGUCAA--UGCCGGGUCACACUA---GUCCCGCACUU-GAC --...........(((..(((.....(((((((---.(..(.(((....)))...)..)((((((((.....))))))))..--.)))))))......)---))...)))...-... ( -26.50, z-score = -0.29, R) >droEre2.scaffold_4690 5518012 107 - 18748788 AAUAUAUACAUAA-GUAUAGGUAUUCACCUGGC---UAGUCAUUGAUUCGAAAUUGGCCGGGUUCCAAUCGCUGGAGGUCAA--AGCCGGGUCACACCA---GUCCCUCACUU-GAC ..........(((-((...(((....(((((((---((((............)))))))))))((((.....))))......--.)))(((.(......---).)))..))))-).. ( -28.60, z-score = -0.86, R) >droVir3.scaffold_12970 5484938 100 - 11907090 ---------AAGAUAUUCUAUCAUUCUCAAA-----UGUUAUGCAAUUUAAACAAAGUACGCUUAUUAUCAUAUAAA---AAAUAGCUGAGGCAAAUUAAUUUUUGUUUAGACAAGC ---------..((((...)))).........-----......((..(((((((((((.....((((......)))).---.....((....))........)))))))))))...)) ( -8.90, z-score = 1.58, R) >consensus ______AAUAUAAUGUAUAAGCAUUCACCUGGC___UGGUCAUUGAUUCCAAUUUGGCAGGCUUCCAAUCGCUGGAGGUCAA__AGCCGGGUCACACCA___GUUCCUCACUU_GAC ..........................(((((((.....((((............)))).((((((((.....)))))))).....)))))))......................... (-15.70 = -16.57 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:06 2011