
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,629,857 – 13,629,954 |
| Length | 97 |
| Max. P | 0.706056 |

| Location | 13,629,857 – 13,629,954 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.94 |
| Shannon entropy | 0.62712 |
| G+C content | 0.57632 |
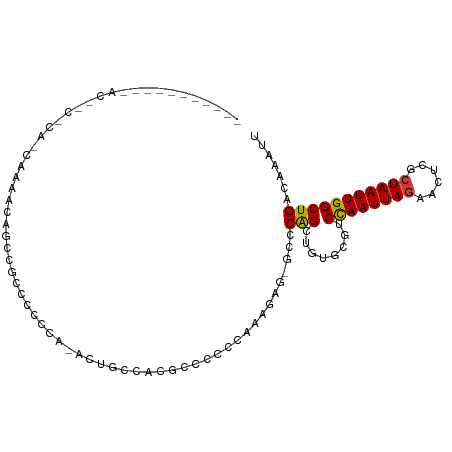
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -7.74 |
| Energy contribution | -7.64 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
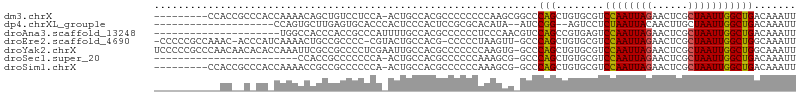
>dm3.chrX 13629857 97 - 22422827 ---------CCACCGCCCACCAAAACAGCUGUCCUCCA-ACUGCCACGCCCCCCCCAAGCGGCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGACAAAUU ---------...............(((((((.......-...(((.(...........).))).))))))).((.((((((((......)))))))).))....... ( -20.80, z-score = -1.34, R) >dp4.chrXL_group1e 1276225 83 - 12523060 --------------------CCAGUGCUUGAGUGCACCCACUCCCACUCCGCGCACAUA--AUCCGG--AGUCCUCUAAUUACAACUUGCUAAUUGGCUGACAAAUU --------------------...((((..(((((..........)))))...))))...--......--.(((..(((((((........)))))))..)))..... ( -17.50, z-score = -1.48, R) >droAna3.scaffold_13248 2851709 86 - 4840945 ---------------------UGGCCACCCACCGCCCAUUUUGCCACGCCCCCCUCCCAACGUCCAGCCGUGAGUCCAAUUAGAACUCGCUAAUUGGCUGACAAAUU ---------------------.(((........))).........................(((.((((((((((.(.....).)))))).....)))))))..... ( -17.90, z-score = -1.66, R) >droEre2.scaffold_4690 5495010 102 + 18748788 -CCCCCGCCAAAC-ACCCAUCAAAACUGCCGCCCC-CGUACUGCCACG-CCCCCUAAGUU-GCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGGCAAAUU -.....((((...-.....................-(((((.((...(-(..........-))...)).))))).((((((((......)))))))).))))..... ( -22.40, z-score = -1.55, R) >droYak2.chrX 7923857 106 - 21770863 UCCCCCGCCCAACAACACACCAAAUUCGCCGCCCCUCGAAUUGCCACGCCCCCCCAAGUG-GCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGGCAAAUU ......(((......((((.(.((((((........))))))(((((..........)))-))...).)))).(.((((((((......))))))))).)))..... ( -27.70, z-score = -2.69, R) >droSec1.super_20 268264 81 - 1148123 ------------------------CCACCGCCCCCCCA-ACUGCCACGCCCCCCAAAGCG-GCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGACAAAUU ------------------------..............-.(((...(((........)))-...))).(((.((.((((((((......)))))))).))))).... ( -16.60, z-score = -0.80, R) >droSim1.chrX 10436360 96 - 17042790 ---------CCACCGCCCACCAAAACCGCCGCCCCCCA-ACUGCCACGCCCCCCAAAGCG-GCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGACAAAUU ---------..................(((((......-...((...))........)))-)).....(((.((.((((((((......)))))))).))))).... ( -18.93, z-score = -0.91, R) >consensus ___________AC__C_CA_CAAAACAGCCGCCCCCCA_ACUGCCACGCCCCCCAAAGAG_GCCCAGCUGUGCGUCCAAUUAGAACUCGCUAAUUGGCUGACAAAUU ................................................................(((........((((((((......)))))))))))....... ( -7.74 = -7.64 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:03 2011