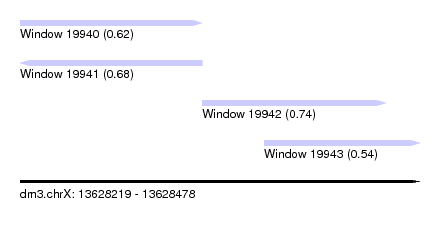
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,628,219 – 13,628,478 |
| Length | 259 |
| Max. P | 0.739309 |

| Location | 13,628,219 – 13,628,337 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 54.31 |
| Shannon entropy | 0.62748 |
| G+C content | 0.50126 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -13.05 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13628219 118 + 22422827 CCACACCCAGCGGUGUGGGCGUGGAACAAAGGUUGCUCCUUCGUUCGCCUUGUUUCCGCCGAUUCAGUUGUUAGCUUAACGGCUGUUGCUGUUUUAGUCCUUAUUGUCGCACUUGCCU .((((((....))))))(((((((((..(((((.((......))..)))))..))))))(((..((((.....(((.((((((....))))))..)))....))))))).....))). ( -38.10, z-score = -1.73, R) >droSim1.chrX 10434695 107 + 17042790 CCACACCCAUCGGCGUGGGCGUGGGACAAAGGU-----------UCGCCUUGUUUCCGCCGAUUCAGUUGUUAGCUUAACGGCUGUUGCUGUUUUAGUCCUUAUUGUCGCACUUGCCU ...........((((((((((.(..((((.((.-----------...))))))..)))))((..((((.....(((.((((((....))))))..)))....)))))).)))..))). ( -33.70, z-score = -0.97, R) >droVir3.scaffold_12970 8005025 92 + 11907090 -----------------CAUCUGGCGCAAGGUUC------UGGUUUUUGUUAAUAGAGCUAAUUCGGA-AUUGUCCAGAGAGCGGAACUUGAUUCAGAUAG-GCUUUCACAUUGGCC- -----------------.((((((..((((.(((------((.((((((.((((.(((....)))...-))))..)))))).)))))))))..)))))).(-(((........))))- ( -25.40, z-score = -1.14, R) >consensus CCACACCCA_CGG_GUGGGCGUGGAACAAAGGU_______U_GUUCGCCUUGUUUCCGCCGAUUCAGUUGUUAGCUUAACGGCUGUUGCUGUUUUAGUCCUUAUUGUCGCACUUGCCU (((((((....)))))))...((((((((((((.............)))))......((((.((.(((.....))).))))))......)))))))...................... (-13.05 = -14.12 + 1.07)


| Location | 13,628,219 – 13,628,337 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 54.31 |
| Shannon entropy | 0.62748 |
| G+C content | 0.50126 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -13.13 |
| Energy contribution | -14.63 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13628219 118 - 22422827 AGGCAAGUGCGACAAUAAGGACUAAAACAGCAACAGCCGUUAAGCUAACAACUGAAUCGGCGGAAACAAGGCGAACGAAGGAGCAACCUUUGUUCCACGCCCACACCGCUGGGUGUGG .(((.(((.(........).)))......((....((((...((.......))....))))(....)...))(((((((((.....)))))))))...)))((((((....)))))). ( -38.60, z-score = -2.20, R) >droSim1.chrX 10434695 107 - 17042790 AGGCAAGUGCGACAAUAAGGACUAAAACAGCAACAGCCGUUAAGCUAACAACUGAAUCGGCGGAAACAAGGCGA-----------ACCUUUGUCCCACGCCCACGCCGAUGGGUGUGG .(((..(((.(((((..(((...............((((...((.......))....))))(....).......-----------.)))))))).))))))((((((....)))))). ( -32.50, z-score = -1.17, R) >droVir3.scaffold_12970 8005025 92 - 11907090 -GGCCAAUGUGAAAGC-CUAUCUGAAUCAAGUUCCGCUCUCUGGACAAU-UCCGAAUUAGCUCUAUUAACAAAAACCA------GAACCUUGCGCCAGAUG----------------- -((((((.((((.(((-....((......))....))).))((((....-))))........................------..)).))).))).....----------------- ( -13.90, z-score = 0.28, R) >consensus AGGCAAGUGCGACAAUAAGGACUAAAACAGCAACAGCCGUUAAGCUAACAACUGAAUCGGCGGAAACAAGGCGAAC_A_______ACCUUUGUCCCACGCCCAC_CCG_UGGGUGUGG ......(((.(((((....................((((...((.......))....))))(....)......................))))).)))..(((((((....))))))) (-13.13 = -14.63 + 1.51)



| Location | 13,628,337 – 13,628,456 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.20 |
| Shannon entropy | 0.75488 |
| G+C content | 0.51931 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.90 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13628337 119 + 22422827 ACAAGGACCCCCCAUAUAUACCGACCGGCCGCCAGGCAAUUUGCAUCAAGUGCAGCCAUGCACUUGGUC-GACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAA ....((.....)).............(((.((((..(((.(((.((((((((((....)))))))))))-))..)))..)))).))).........(((((((......))))).))... ( -40.60, z-score = -2.48, R) >droSim1.chrX 10434802 115 + 17042790 ACAAGGACCC--CAUGUAUACCGCCCGGCC--CAGGCAAUUUGCAUCAAGUGCAGCCGUGCACUUGGUC-GACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAA (((.((...)--).))).....((..((((--((..(((.(((.((((((((((....)))))))))))-))..)))..)))..)))...........(((((......)))))..)).. ( -34.80, z-score = -0.08, R) >droSec1.super_20 266727 115 + 1148123 ACAAGGACCC--CAUAUACACCGCCCGGCC--CAGGCAAUUUGCAUCAAGUGCAGCCGUGCACUUGGUC-GACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAA ..........--..........((..((((--((..(((.(((.((((((((((....)))))))))))-))..)))..)))..)))...........(((((......)))))..)).. ( -33.80, z-score = -0.23, R) >droYak2.chrX 7922362 108 + 21770863 ACAACUACCC---CCAUACCGCCGCCAGCU--CAGGCAAUUUGCAUCAAGUGCAGCCAGGCACUCGCUC-GACUUUGAAUGGCUGCCACCAGAAUCCGGUGCA------UGCACACGCAG ..........---.......((.(((....--..)))....(((((.....(((((((..((.((....-))...))..)))))))((((.......)))).)------))))...)).. ( -29.30, z-score = -0.18, R) >droEre2.scaffold_4690 5493418 104 - 18748788 ACAAGUACAC---C-AUACCUCC---AGCU--GAGGCAAUUUGCAUCAAGUGCAGCCAGGCACUUGGUC-GACUUUGAAUGGCUGCCACCAAAAUGCUGUGCA------UGCACACGCAA ....(((((.---.-...((((.---....--))))......((((...(((((((((..((...((..-..)).))..)))))).)))....))))))))).------(((....))). ( -30.00, z-score = 0.15, R) >droAna3.scaffold_13248 2850188 91 + 4840945 ACAAGGACCU---CCCUACCCC-----------AGGCAAUUUGCGUCAAGUGCAAAAAGGCAUUUCGCCUGGCUUUCAGUGGC---AACUGGAAGCCGAAACAAGAAA------------ ...(((....---.))).....-----------......((((((.....)))))).((((.....))))((((((((((...---.))))))))))...........------------ ( -27.50, z-score = -1.14, R) >droVir3.scaffold_12970 8005117 91 + 11907090 ---------------------GGAAUAGCUGUGCGAACAUUUGCAAUAAGUGAAAUCUUCUUCGUAGAC--AUCAUUAAAGGU-AUACCAAGAAACCAAUAGGGUUACAUUUAAG----- ---------------------((.(((.((.(((((....)))))...(((((..(((.......))).--.)))))..)).)-)).))..(.((((.....)))).).......----- ( -15.60, z-score = -0.72, R) >consensus ACAAGGACCC___AUAUACACCG_CCAGCC__CAGGCAAUUUGCAUCAAGUGCAGCCAUGCACUUGGUC_GACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAA .........................(((((.....((.....))(((((((((......)))))))))............))))).............(((((......)))))...... (-13.55 = -14.90 + 1.35)
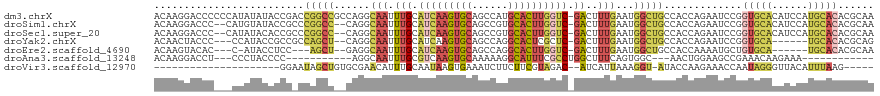

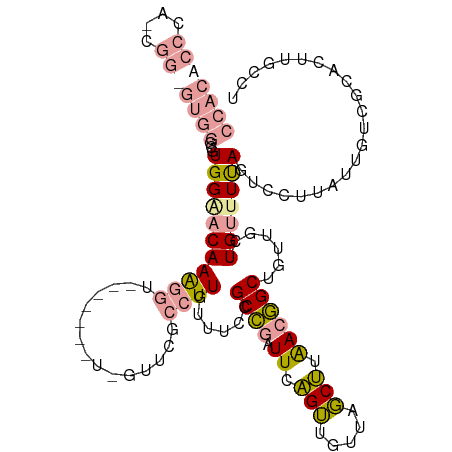
| Location | 13,628,377 – 13,628,478 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.60 |
| Shannon entropy | 0.54322 |
| G+C content | 0.49398 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -17.22 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13628377 101 + 22422827 UUGCAUCAAGUGCAGCCAUGCACUUGGUCGACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAAUCAUCUGCACUGAUGAUUCUAA------------------ (((.((((((((((....)))))))))))))........((......))(((((((((((((.((...(((....)))...)).)))))))..))))))..------------------ ( -32.60, z-score = -1.86, R) >droSim1.chrX 10434838 119 + 17042790 UUGCAUCAAGUGCAGCCGUGCACUUGGUCGACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAAUCAUCUGCACUGCUGAUUCUAAUGCUAAGCCGUCGUCUUG ..(.((((((((((....)))))))))))(((...((.(((((..((..(((((((((((((.((...(((....)))...)).)))))))..))))))..))...))))))))))... ( -40.30, z-score = -2.08, R) >droSec1.super_20 266763 119 + 1148123 UUGCAUCAAGUGCAGCCGUGCACUUGGUCGACUUUGAAUGGCUGCCACCAGAAUCCGGUGCACAUCCAUGCACACGCAAUCAUCUGCACUGCUGAUUCUAAUGCUAAGCCGUCGUCUUG ..(.((((((((((....)))))))))))(((...((.(((((..((..(((((((((((((.((...(((....)))...)).)))))))..))))))..))...))))))))))... ( -40.30, z-score = -2.08, R) >droYak2.chrX 7922397 111 + 21770863 UUGCAUCAAGUGCAGCCAGGCACUCGCUCGACUUUGAAUGGCUGCCACCAGAAUCCGGUGCA------UGCACACGCAGUCAUCUGCACAGCUGCAUCUAACGCCAAGCCGUCGUCU-- ..((.((..(((((((((..((.((....))...))..)))))).)))..))....((((((------.((....((((....))))...))))))))....)).............-- ( -32.10, z-score = 0.55, R) >droEre2.scaffold_4690 5493449 111 - 18748788 UUGCAUCAAGUGCAGCCAGGCACUUGGUCGACUUUGAAUGGCUGCCACCAAAAUGCUGUGCA------UGCACACGCAAUCAUCUGCACUGCUUAAUCUAACGCCAAGCCGUCGUCC-- (((.(((((((((......))))))))))))....((.(((((((.........((.(((((------(((....)))......))))).))..........))..)))))))....-- ( -30.81, z-score = -0.04, R) >droVir3.scaffold_12970 8005136 98 + 11907090 UUGCAAUAAGUGAAAUCUUCUUCGUAGACAUCAUUAAAGGUAUACCA--AGAAACCAAUAGG------GUUACAUUUAAGUGAUU-UAUGGAUGGAUACCAACCCAU------------ ........(((((..(((.......)))..)))))...(((((.(((--.....(((...((------(((((......))))))-).))).)))))))).......------------ ( -17.30, z-score = -0.25, R) >consensus UUGCAUCAAGUGCAGCCAUGCACUUGGUCGACUUUGAAUGGCUGCCACCAGAAUCCGGUGCA______UGCACACGCAAUCAUCUGCACUGCUGAAUCUAACGCCAAGCCGUCGUCU__ ((((.....(.((((((((.((.((....))...)).)))))))))...........(((((......)))))..))))........................................ (-17.22 = -16.62 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:41:02 2011