
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,622,021 – 13,622,143 |
| Length | 122 |
| Max. P | 0.704992 |

| Location | 13,622,021 – 13,622,115 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.47 |
| Shannon entropy | 0.71779 |
| G+C content | 0.38395 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -7.81 |
| Energy contribution | -7.56 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
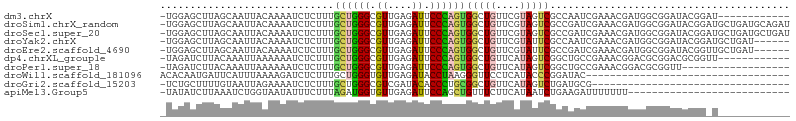
>dm3.chrX 13622021 94 + 22422827 -GUAAUAAUGAUUAAUUAGAUUAGUGGAACUUG-GAGCUUA----GCAAUU---ACAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUC -........(((((....((.((((((((((..-(.(((((----((((..---........--------...))))))))).)..)).)))).....)))).)).))))) ( -24.42, z-score = -1.54, R) >droSim1.chrX_random 3696043 94 + 5698898 -GUAAUAAUGAUUAAUUAUAAUAGUGGAACUUG-GAGCUUA----GCAAUU---ACAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUC -.............(((((((((((((((((..-(.(((((----((((..---........--------...))))))))).)..)).)))).....)))))).))))). ( -23.92, z-score = -1.68, R) >droSec1.super_20 260335 94 + 1148123 -GUAAUAAUGAUUAAUUAUAAUAGUGGAACUUG-GAGCUUA----GCAAUU---ACAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUC -.............(((((((((((((((((..-(.(((((----((((..---........--------...))))))))).)..)).)))).....)))))).))))). ( -23.92, z-score = -1.68, R) >droYak2.chrX 7916035 94 + 21770863 -GUAAUAAUGAUUAAUUAUAAUAGUGGAACUUG-GAGCUUA----GCAAUU---ACAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAUUC -...(((((.....)))))((((((((((((..-(.(((((----((((..---........--------...))))))))).)..)).)))).....))))))....... ( -22.62, z-score = -1.44, R) >droEre2.scaffold_4690 5487000 94 - 18748788 -GUAAUAAUGGUUAAUUAUAAUAGUGGAACUUG-GAGCUUA----GCAAUU---ACAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAUUC -...(((((.....)))))((((((((((((..-(.(((((----((((..---........--------...))))))))).)..)).)))).....))))))....... ( -22.62, z-score = -1.25, R) >droAna3.scaffold_13248 2844208 97 + 4840945 UGUA-UAAUUAGUUAUAAUAUUAGUGGAACUUG-GAGCUUACUUAGAAAUU---AAAAAAUC--------UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAUU- .(((-(...(((((((.........((((((..-(.((((((..(((.(((---....))).--------)))..).))))).)..)).))))..)))))))...)))).- ( -17.10, z-score = 0.36, R) >droVir3.scaffold_12970 7998984 84 + 11907090 -----------AUGAAGAUGCGUGUUCUAUUCCAUUAUUUAG----UACCU---AGAAAAUC--------UCUUUGCUGGGCGUCGAAAUGCCCAUGGGCUGUUCGUAGU- -----------.....((((((((........)))...((((----((...---(((.....--------))).)))))))))))(((..(((....)))..))).....- ( -17.50, z-score = 0.06, R) >droMoj3.scaffold_6482 1203460 79 - 2735782 -------------AUUGAUGCAUGUC---UCUGGCUAGCUAC----UAUCU---AGAAAAUC--------UCCUUGCUGGGCGUCGAAAUGCCCAUCGGCUGCUCGUACU- -------------...((.(((.(((---(((((.(((...)----)).))---))).....--------.....(.(((((((....))))))).))))))))).....- ( -21.80, z-score = -1.63, R) >droGri2.scaffold_15203 923848 85 - 11997470 -----------ACGAUGAUGCAUUGAUGUUUUGUCUGCUUUUG---UAAUU---AGAAAAUC--------UCUUUGCUGGGCGUCGAUACACCCUGCGGCUGUUCAUAGU- -----------...((((.((...(((.(((((..(((....)---))..)---)))).)))--------....(((.(((.((....)).))).)))))...))))...- ( -18.40, z-score = -0.15, R) >apiMel3.Group5 1755659 99 - 13386189 -GUUCUAAAAUUGUAUUAUCCU--CAGAAUUUGUAAAAUUAUAUCUUAAAUCUGGUAAUAUU--------UCUUUAGAUGGUGUUGAGAUUCCAGCUGUUUCUUCAUAAU- -..((((((...(((((((...--((((.((((.............))))))))))))))).--------..))))))..(((..(((((.......)))))..)))...- ( -14.02, z-score = -0.47, R) >triCas2.ChLG6 4181592 100 + 13544221 ----AUUAAUUUGAAUGAUUAUAAAAUGAUUUGUAGGCGCACA---UAAAG---AGAAAAUCAUUGGAAUUCCUUGCUGGGUGUUGAAAUACCUAGCGCUUCCUCGUAGC- ----...(((((.(((((((........((.(((....))).)---)....---....))))))).)))))....(((((((((....)))))))))(((.......)))- ( -21.89, z-score = -1.10, R) >consensus _GUAAUAAUGAUUAAUUAUAAUAGUGGAACUUG_GAGCUUA____GCAAUU___ACAAAAUC________UCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGU_ ..........................................................................(((((((.((....)).)))))))............. ( -7.81 = -7.56 + -0.25)



| Location | 13,622,051 – 13,622,143 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 64.83 |
| Shannon entropy | 0.69143 |
| G+C content | 0.45587 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -8.89 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13622051 92 + 22422827 -UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUCGCCAAUCGAAACGAUGGCGGAUACGGAU------------ -....(((((((((.............)))))))))...................(((((((..(((((.(((...))))))))..)))))))------------ ( -29.22, z-score = -1.43, R) >droSim1.chrX_random 3696073 104 + 5698898 -UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUCGCCGAUCGAAACGAUGGCGGAUACGGAUGCUGAUGCAGAU -....(((((((...............(..(((((.((....)).)))))..)..(((((((..((((.((((...))))))))..)))))))))))).)).... ( -36.80, z-score = -2.06, R) >droSec1.super_20 260365 104 + 1148123 -UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAGUCGCCGAUCGAAACGAUGGCGGAUACGGAUGCUGAUGCUGAU -...((((((((...............(..(((((.((....)).)))))..)..(((((((..((((.((((...))))))))..)))))))))))).)))... ( -37.60, z-score = -2.41, R) >droYak2.chrX 7916065 98 + 21770863 -UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAUUCGCCAAUCGAAACGAUGGCGGAUACGGAUGCUGAU------ -......(((((...............(..(((((.((....)).)))))..)..((((((((((((((.(((...)))))))))))))))))))))).------ ( -37.20, z-score = -3.46, R) >droEre2.scaffold_4690 5487030 98 - 18748788 -UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCGUAUUCGCCGAUCGAAACGAUGGCGGAUACGGUUGCUGAU------ -....(((((((((.............)))))))))...........(((..((....((((((((((.((((...))))))))))))))))..)))..------ ( -37.52, z-score = -3.26, R) >dp4.chrXL_group1e 1266233 92 + 12523060 -UAGAUCUUACAAAUUAAAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCAUAGUCGGCUGCCGAAACGGACGCGGACGCGGUU------------ -............................((((((.((....)).))))))((((((.....(((.((..(((...)))..)).)))))))))------------ ( -22.00, z-score = 0.76, R) >droPer1.super_18 112394 86 + 1952607 -UAGAUCUUACAAAUUAAAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCAUAGUCGGCUGCCGAAACGGACGCGGUU------------------ -..((((......................((((((.((....)).)))))).((.((((....((((...))))...))))))))))------------------ ( -18.60, z-score = 0.76, R) >droWil1.scaffold_181096 7676966 71 + 12416693 ACACAAUGAUUCAUUUAAAAGAUCUCUUUGCUGGGUGUUGAGAUACCUAAGGGUUCCUCAUACCCGGAUAC---------------------------------- .................((((....)))).((((((((.(((..(((....)))..)))))))))))....---------------------------------- ( -20.40, z-score = -2.37, R) >droGri2.scaffold_15203 923870 71 - 11997470 -UCUGCUUUUGUAAUUAGAAAAUCUCUUUGCUGGGCGUCGAUACACCCUGCGGCUGUUCAUAGUCUGAUGCG--------------------------------- -...(((((((....))))).(((.....((.(((.((....)).))).))(((((....))))).))))).--------------------------------- ( -13.60, z-score = 0.30, R) >apiMel3.Group5 1755695 77 - 13386189 -UAUAUCUUAAAUCUGGUAAUAUUUCUUUAGAUGGUGUUGAGAUUCCAGCUGUUUCUUCAUAAUCUGAAGAUUUUUUU--------------------------- -..((((........)))).....(((((((((.(((..(((((.......)))))..))).))))))))).......--------------------------- ( -16.60, z-score = -2.11, R) >consensus _UGGAGCUUAGCAAUUACAAAAUCUCUUUGCUGGGCGUUGAGAUUCCCAGUGGCUGUUCAUAGUCGCCUAUCGAAACGAUGGCGGAUACGG_U____________ .............................((((((.((....)).))))))(((((....)))))........................................ ( -8.89 = -9.02 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:59 2011