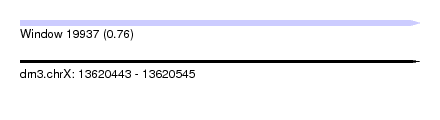
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,620,443 – 13,620,545 |
| Length | 102 |
| Max. P | 0.755191 |

| Location | 13,620,443 – 13,620,545 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.40 |
| Shannon entropy | 0.75081 |
| G+C content | 0.61409 |
| Mean single sequence MFE | -39.41 |
| Consensus MFE | -12.54 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
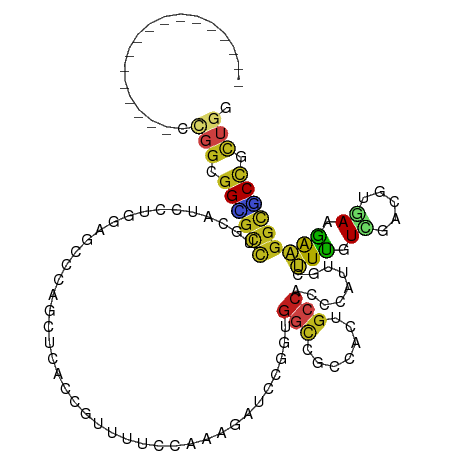
>dm3.chrX 13620443 102 + 22422827 -----------------CCGGCGGCGCUUGCAUCCUGGAGCCAACAACUCCGUUCUCCAAAGAUCCUGUGGCCAAAAGUGCCAGCCAUUGUUUUGUCGAUGUGAAGAAGGCGCCGCUGU -----------------.((((((((((((((((.(((((..(((......))))))))(((((...(((((...........))))).)))))...))))).....))))))))))). ( -37.10, z-score = -1.84, R) >droSec1.super_20 258805 102 + 1148123 -----------------CCGGCGGCGCCUGCAUCCUGGAGCCCACGACUCCGUUCUCCAAAGAUCCUGUGGCCAAAAGUGCCAGCCAUUGUUUUGUCGAUGUGAAGAAGGCGCCGUUGU -----------------.((((((((((((((((.(((((...(((....))).)))))(((((...(((((...........))))).)))))...))))).....))))))))))). ( -38.50, z-score = -2.31, R) >droYak2.chrX 7914579 102 + 21770863 -----------------CCGGCGGCGCCUGCAUCCUGGAGCCCAGCACGCCGUUUUCCAAAGAUCCAGUGGCCAAAAGUGCCACCCAUUGCUUUGUCGAUGUGAAGAAGGCGCCGCUGU -----------------.(((((((((((.(...((((...))))((((.((.....(((((.....((((((....).)))))......))))).)).))))..).))))))))))). ( -42.20, z-score = -2.73, R) >droEre2.scaffold_4690 5485427 102 - 18748788 -----------------CCGGCGGCGCCUGCAUCCUGGAGCCCAGCACGCCGUUUUCCAAAGAUCCAGUGGCCAAAAGUGCCACCCAUUGCUUUGUCGAUGUGAAGAAGGCGCCGCUGU -----------------.(((((((((((.(...((((...))))((((.((.....(((((.....((((((....).)))))......))))).)).))))..).))))))))))). ( -42.20, z-score = -2.73, R) >droAna3.scaffold_13248 2842481 102 + 4840945 -----------------CCGGCGGAGCCGUCAUCCUGGAGCCCAAGUCGCCCUUUUCGGCGGAUCCCAUUGCCGCCACCGCCACCCACUGCUUCGUGGACGUGAAGAAGGCUCCGCUCU -----------------..(((((((((.((((((((((((....((.((.......(((((.........)))))...)).)).....)))))).)))..)))....))))))))).. ( -39.30, z-score = -1.22, R) >dp4.chrXL_group1e 1262965 102 + 12523060 -----------------CUGGCGGGGCCUGCAUUUUAGAGCCGCGCUCUCCGUUUGCCCAAGAUCCGCUGGUGGCCACAGCCACCGAUUGUUUUGUGGACAUCAAGAAGGCCCCGCUGG -----------------(..((((((((((((....(((((...))))).....)))....((((((((((((((....)))))))........)))))..))....)))))))))..) ( -47.60, z-score = -3.15, R) >droPer1.super_18 109740 102 + 1952607 -----------------CUGGCGGGGCCUGCAUUUUAGAGCCGCGCUCUCCGUUUGCCCAAGAUCCGCUGGUGGCCACAGCCACCGAUUGUUUUGUGGACAUCAAGAAGGCCCCGCUGG -----------------(..((((((((((((....(((((...))))).....)))....((((((((((((((....)))))))........)))))..))....)))))))))..) ( -47.60, z-score = -3.15, R) >droWil1.scaffold_180777 4498698 102 + 4753960 -----------------CCGGCGGAGCUGCAAUCGUACAGCCACAUUCACCCUUUGCCGAUGAUCCGGUGGCCGGCACAGCCACUCAUUGCUUUGUCGAUAUGAAGAAGGCCCCCCUGG -----------------((((.((.(((....(((((..(((((..(((.(.......).)))....)))))(((((.(((........))).))))).)))))....))).)).)))) ( -31.80, z-score = -0.27, R) >droVir3.scaffold_12970 601775 102 - 11907090 -----------------CCGGCGGUGCCACCAUUUUGGAGCCAAGUUCACCCUUCAGCUCGGACGCAACGGCGGCGAGUGCCACCCACUGUUUCGUAGACAUCAAGAAGGCGCCUCUGG -----------------((((.((((((....(((((((((.(((......)))..)))).((.(((..((.(((....))).))...))).))........))))).)))))).)))) ( -33.50, z-score = -0.05, R) >droMoj3.scaffold_6473 10009617 102 + 16943266 -----------------CUGGCGGCGCCACGAUCCUGGAGCCCAGCUCACCCUUCAGCGCGGACGCCACGUCAGCCACUGCCACCCACUGCUUCGUCGACGCCAAGAAGGCGCCGCUGG -----------------(..((((((((.((((...(((((...(((........)))((.((((...)))).))..............))))))))).(.....)..))))))))..) ( -37.20, z-score = -0.13, R) >droGri2.scaffold_15203 8719035 102 - 11997470 -----------------CCGGCGGUGCCCAUAUUUUGGAGCCCAGCACACCCUUCAGCUCGGAUGCGUCGGCGGCGAGGGCCACACACUGCUUCGUCGAUGCGAAGAAGGCACCGCUGG -----------------(((((((((((......((.((((..((......))...)))).))(((((((((((.(.((........)).).))))))))))).....))))))))))) ( -45.80, z-score = -2.04, R) >anoGam1.chr2R 26787083 99 + 62725911 -----------------CCGGCGGCGGUUACGUGCUGGACGUGGAUCCACCGUUCAUCCACUCGGAGCGGGCGAUGGCGGCCACUCACUGCUUCGUCGACCGGAAGGCGAAACUGU--- -----------------((((((.((....))))))))..((((((.........)))))).((((((((..(((((...))).)).))))))))(((.((....)))))......--- ( -38.40, z-score = 0.21, R) >triCas2.ChLG2 3502433 114 - 12900155 UGGCGUAAGUUGAAAUUUUGGCAGGGUUUUGGGUUUUACGCGGGGUUUUAGGCCUCC---GUUUAAGGAGAACGUUCACGCCACUCACUGCCUUUUGGAAAUUAAAAGUGUCCCCGA-- ..((((((((..((((((.....))))))..)..)))))))((((.....(((((((---......))))...(....))))...((((.((....))........)))).))))..-- ( -31.10, z-score = 0.54, R) >consensus _________________CCGGCGGCGCCUGCAUCCUGGAGCCCAGCUCACCGUUUUCCAAAGAUCCGGUGGCCGCCACUGCCACCCAUUGCUUUGUCGACGUGAAGAAGGCGCCGCUGG ..................(((.((((((.........................................(((.......))).........(((.((.....)).))))))))).))). (-12.54 = -12.14 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:58 2011