
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,614,402 – 13,614,473 |
| Length | 71 |
| Max. P | 0.514464 |

| Location | 13,614,402 – 13,614,473 |
|---|---|
| Length | 71 |
| Sequences | 11 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 50.88 |
| Shannon entropy | 1.07136 |
| G+C content | 0.46414 |
| Mean single sequence MFE | -14.05 |
| Consensus MFE | -4.99 |
| Energy contribution | -5.20 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
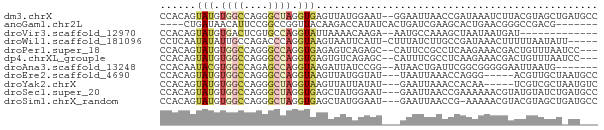
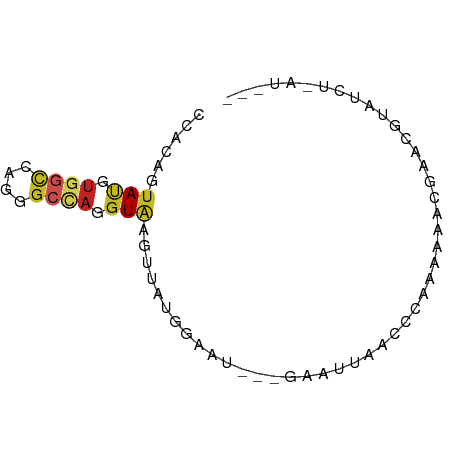
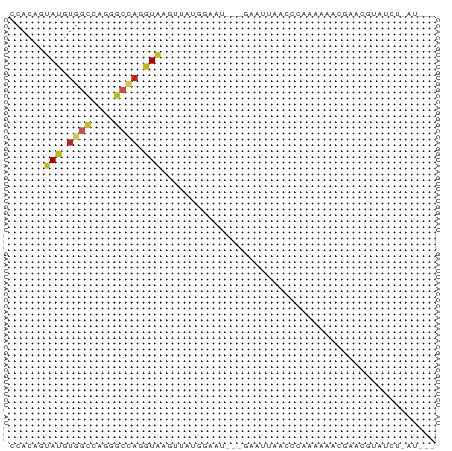
>dm3.chrX 13614402 71 - 22422827 CCACAGUAUGUGGCCAGGGCUAGGUGAGUUAUGGAAU--GGAAUUAACCGAUAAAUCUUACGUAGCUGAUGCC ((((.....))))....(((((.(((((........(--((......)))......))))).)))))...... ( -17.84, z-score = -0.76, R) >anoGam1.chr2L 13101304 62 - 48795086 ----CUGAUAACAUUCCGGCCGGUUACAAGACCAUAUCACUGAUCGAAGCACUGAACGGGCCGACG------- ----............(((((((((....))))...(((.((.......)).)))...)))))...------- ( -15.50, z-score = -1.70, R) >droVir3.scaffold_12970 11636016 58 + 11907090 CCACAGUAUGUGACUCGUGCCAGGUAUUAAAACAAGA--AAUGCCAAAGCUAAUAAUGAU------------- .....(((((.....)))))..((((((.........--))))))...............------------- ( -8.00, z-score = -0.40, R) >droWil1.scaffold_181096 7667191 67 - 12416693 CCUCAAUAUAUUGCCAGACCCAGGUAAGUAAUUCAUU-CUUUAUCUUGCCGAUAAACUUUUUAAUAUU----- ......................(((((((((......-..))).))))))..................----- ( -7.80, z-score = -1.14, R) >droPer1.super_18 103988 68 - 1952607 CCACAGUAUGUGGCCAGGGCCAGGUGAGAGUCAGAGC--CAUUCCGCCUCAAGAAACGACUGUUUAAUCC--- ..(((((.((((((....)))(((((.((((......--.)))))))))......)))))))).......--- ( -18.50, z-score = -1.00, R) >dp4.chrXL_group1e 1257213 68 - 12523060 CCACAGUAUGUGGCCAGGGCCAGGUGAGUGUCAGAGC--CAUUUCGCCUCAAGAAACGACUGUUUAAUCC--- ..(((((.((((((....)))(((((((((.......--)).)))))))......)))))))).......--- ( -18.20, z-score = -0.76, R) >droAna3.scaffold_13248 2836426 64 - 4840945 CCACAAUACGUGGCCAGAGCCAGGUAAGAUUAUCCGG--AUAACUGAUUCGGCGGGGGAAUUAAUG------- ((....(((.((((....)))).))).......((((--((.....)))))).))...........------- ( -16.40, z-score = -0.89, R) >droEre2.scaffold_4690 5479355 65 + 18748788 CCACAGUAUGUGGCCAGGGCCAGGUAAGUUAUGGUAU---UAAUUAAACCAGGG-----ACGUUGCUAAUGCC .....((((.((((....))))((((((((.((((..---.......))))..)-----)).))))).)))). ( -16.70, z-score = -0.07, R) >droYak2.chrX 7908495 65 - 21770863 CCACAGUAUGUGGCCAGGGCUAGGUAAGUUAUUAUAU---GAAUUAAACCACAA-----UCGUCGCUAAUGUC ......(((.((((....)))).)))...........---..............-----.............. ( -7.40, z-score = 1.75, R) >droSec1.super_20 252754 70 - 1148123 CCACAGUAUGUGGCCAGGGCUAGGUGAGCUAUGGAAU---GAAUUAACCGAAAAAACGUAUGUAUCUGAUGCC ((((.....))))(((.((((.....)))).)))...---.................((((.......)))). ( -12.30, z-score = 0.64, R) >droSim1.chrX_random 3690938 69 - 5698898 CCACAGUAUGUGGCCAGGGCUAGGUGAGCUAUGGAAU---GAAUUAACCG-AAAAACGUACGUAGCUGAUGCC ((((.....))))....(((...((.(((((((..((---(.........-.....))).))))))).))))) ( -15.94, z-score = -0.36, R) >consensus CCACAGUAUGUGGCCAGGGCCAGGUAAGUUAUGGAAU___GAAUUAACCCAAAAAACGAACGUAUCU_AU___ ......(((.((((....)))).)))............................................... ( -4.99 = -5.20 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:56 2011