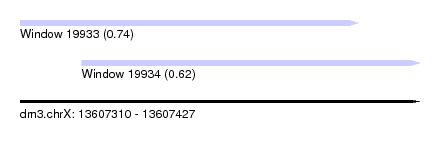
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,607,310 – 13,607,427 |
| Length | 117 |
| Max. P | 0.738480 |

| Location | 13,607,310 – 13,607,409 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.71388 |
| G+C content | 0.51330 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.24 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
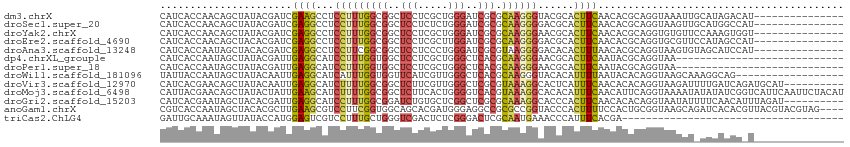
>dm3.chrX 13607310 99 + 22422827 CAUCACCAACAGCUAUACGAUCGAAGCCUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGUACGCACUUCAACACGCAGGUAAAUUGCAUAGACAU--------------- ...........(((((......((((...((((((((((..((.....))..)))).))))))......)))).....((((.....)))))))).)..--------------- ( -25.90, z-score = -0.57, R) >droSec1.super_20 245668 99 + 1148123 CAUCACCAACAGCUAUACGAUCGAGGCCUCCUUUGGCGGCUCCUCUCUGGGAUCGCGCAAGGGGACGCACUUCAACACGCAGGUAAGUUGCAUGGCCAU--------------- ........................(((((((((((((((..((.....))..)))).)))))))..((((((..((......))))).)))..))))..--------------- ( -30.10, z-score = -0.47, R) >droYak2.chrX 7901527 99 + 21770863 CAUCACCAACAGCUAUACGAUCGAGGCCUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGAACGCACUUCAACACGCAGGUGUGUUCCAAAGUGGU--------------- .(((((.....((....(((((..((((.((...)).))))((.....))))))).))...(((((((((((........)))))))))))...)))))--------------- ( -34.10, z-score = -1.24, R) >droEre2.scaffold_4690 5472271 99 - 18748788 CAUCACCAACAGCUACACGAUCGAGGCCUCCUUUGGCGGCUCCUCGCUUGGAUCGCGCAAGGGGACGCACUUCAACACGCAGGUGCGUUCCAUAGCCAU--------------- ...........((((..(((((...(((......)))(((.....)))..)))))(....)(((((((((((........))))))))))).))))...--------------- ( -34.00, z-score = -1.82, R) >droAna3.scaffold_13248 2828917 99 + 4840945 CAUCACCAAUAGCUACACGAUCGAGGCCUCCUUCGGCGGCUCCUCCCUGGGAUCGCGUAAGGGGACACACUUUAACACGCAGGUAAGUGUAGCAUCCAU--------------- ...........(((((((((((..((((.((...)).))))((.....))))))((((.((((......))))...))))......)))))))......--------------- ( -31.00, z-score = -1.55, R) >dp4.chrXL_group1e 1249288 86 + 12523060 CAUCACCAAUAGCUAUACGAUUGAGGCAUCCUUUGGUGGCUCCUCGCUGGGCUCACGCAAGGGAACGCACUUCAAUACGCAGGUAA---------------------------- ...........(((...(((((((((..((((((((((((.((.....)))).))).))))))).....))))))).))..)))..---------------------------- ( -25.20, z-score = -1.40, R) >droPer1.super_18 96135 86 + 1952607 CAUCACCAAUAGCUAUACGAUUGAGGCAUCCUUUGGUGGCUCCUCGCUGGGCUCACGCAAGGGAACGCACUUCAAUACGCAGGUAA---------------------------- ...........(((...(((((((((..((((((((((((.((.....)))).))).))))))).....))))))).))..)))..---------------------------- ( -25.20, z-score = -1.40, R) >droWil1.scaffold_181096 7659733 96 + 12416693 UAUUACCAAUAGCUAUACAAUUGAGGCAUCAUUUGGUGGUUCAUCGUUGGGCUCACGCAAGGGUACACAUUUUAAUACACAGGUAAGCAAAGGCAG------------------ ..(((((...((((..((...((((.((((....)))).))))..))..))))..(....)....................)))))((....))..------------------ ( -20.00, z-score = -1.49, R) >droVir3.scaffold_12970 53411 104 + 11907090 CAUCACGAACAGCUAUACAAUUGAGGCAUCUUUUGGCGGCUCUUCGUUGGGCUCGCGUAAAGGCACUCAUUUCAACACACAGGUAAGAUUUUGAUCAGAUGCAU---------- ((((.................((((((..((((..(((((((......)))).)))..)))))).))))..........((((......))))....))))...---------- ( -21.70, z-score = -0.15, R) >droMoj3.scaffold_6498 423832 114 - 3408170 CAUUACGAACAGCUAUACUAUUGAAGCAUCUUUUGGCGGCUCUUCACUGGGGUCACGUAAAGGCACACAUUUCAACAUUCAGGUAAAAUAUAUAUCGGUCAUUCAAUUCUACAU ......(((..(((......((((((...(((((((.((((((.....)))))).).))))))......))))))......((((.......)))))))..))).......... ( -15.90, z-score = -0.50, R) >droGri2.scaffold_15203 8949324 104 + 11997470 CAUCACGAAUAGCUACACGAUUGAGGCAUCCUUUGGCGGAUCUGUGCUCGGCUCGCGCAAAGGCACCCACUUCAACACACAGGUAAUAUUUUCAACAUUUAGAU---------- ......(((((..(((....((((((...((((((((((..(((....))).)))).))))))......)))))).......))).))))).............---------- ( -18.90, z-score = 0.56, R) >anoGam1.chrX 20338985 110 - 22145176 CGUCACCAAUAGCUACACGCUUGAAGCGUCCUUCGGUGGCAGCACGAUGGGAGGCCGCGCCGGUACCCACUUUUCCACUGCGGUAAGCAGAUCACACGUUACGUACGUAG---- .((((((...(((.....))).((((....)))))))))).....((((((..((((...)))).))).........((((.....)))))))..((((.....))))..---- ( -32.50, z-score = 0.30, R) >triCas2.ChLG4 5843695 77 + 13894384 GAUUGCAAAUAGUUAUACCAUGGAGUCGUCCUUUGCUGGGUCGACUCUCGGGACUCGCAAUGAAACCCAUUUCACGA------------------------------------- .(((((....((((...((..(((((((.(((.....))).))))))).)))))).)))))((((....))))....------------------------------------- ( -21.60, z-score = -1.43, R) >consensus CAUCACCAAUAGCUAUACGAUUGAGGCAUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGAACGCACUUCAACACGCAGGUAAGUU_CAUAGCC_U_______________ ......................((((...(((((((((..(((.....)))..))).))))))......))))......................................... (-11.45 = -11.24 + -0.21)


| Location | 13,607,328 – 13,607,427 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.27 |
| Shannon entropy | 0.75648 |
| G+C content | 0.53215 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -11.47 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
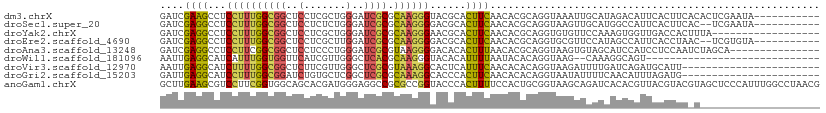
>dm3.chrX 13607328 99 + 22422827 GAUCGAAGCCUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGUACGCACUUCAACACGCAGGUAAAUUGCAUAGACAUUCACUUCACACUCGAAUA----------- ..((((.((..((((((((((..((.....))..)))).))))))...))..........((((.....))))...................))))...----------- ( -27.10, z-score = -1.26, R) >droSec1.super_20 245686 97 + 1148123 GAUCGAGGCCUCCUUUGGCGGCUCCUCUCUGGGAUCGCGCAAGGGGACGCACUUCAACACGCAGGUAAGUUGCAUGGCCAUUCACUUCAC--UCGAAUA----------- ..(((((((.(((((((((((..((.....))..)))).)))))))..)).......((.((((.....)))).)).............)--))))...----------- ( -30.70, z-score = -0.85, R) >droYak2.chrX 7901545 92 + 21770863 GAUCGAGGCCUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGAACGCACUUCAACACGCAGGUGUGUUCCAAAGUGGUUGACCACUUUA------------------ ((((..((((.((...)).))))((.....)))))).(....)(((((((((((........)))))))))))(((((((....))))))).------------------ ( -37.40, z-score = -2.36, R) >droEre2.scaffold_4690 5472289 97 - 18748788 GAUCGAGGCCUCCUUUGGCGGCUCCUCGCUUGGAUCGCGCAAGGGGACGCACUUCAACACGCAGGUGCGUUCCAUAGCCAUUCACCUAAC--UCGUGUA----------- ...(((((((......)))((((((......)))...(....)(((((((((((........)))))))))))...)))..........)--)))....----------- ( -35.80, z-score = -1.91, R) >droAna3.scaffold_13248 2828935 95 + 4840945 GAUCGAGGCCUCCUUCGGCGGCUCCUCCCUGGGAUCGCGUAAGGGGACACACUUUAACACGCAGGUAAGUGUAGCAUCCAUCCUCCAAUCUAGCA--------------- (((.((((.((((((..((((..((.....))..))))..))))))..((((((...(.....)..)))))).........))))..))).....--------------- ( -27.20, z-score = 0.03, R) >droWil1.scaffold_181096 7659751 79 + 12416693 AAUUGAGGCAUCAUUUGGUGGUUCAUCGUUGGGCUCACGCAAGGGUACACAUUUUAAUACACAGGUAAG--CAAAGGCAGU----------------------------- ...((((.((((....)))).))))..(((..(((..(....)..............(((....)))))--)...)))...----------------------------- ( -13.90, z-score = -0.23, R) >droVir3.scaffold_12970 53429 87 + 11907090 AAUUGAGGCAUCUUUUGGCGGCUCUUCGUUGGGCUCGCGUAAAGGCACUCAUUUCAACACACAGGUAAGAUUUUGAUCAGAUGCAUU----------------------- .......((((((....(((((((......)))).))).......................((((......))))...))))))...----------------------- ( -19.20, z-score = -0.18, R) >droGri2.scaffold_15203 8949342 87 + 11997470 GAUUGAGGCAUCCUUUGGCGGAUCUGUGCUCGGCUCGCGCAAAGGCACCCACUUCAACACACAGGUAAUAUUUUCAACAUUUAGAUG----------------------- ..((((((...((((((((((..(((....))).)))).))))))...))((((........)))).......))))..........----------------------- ( -17.20, z-score = 0.45, R) >anoGam1.chrX 20339003 110 - 22145176 GCUUGAAGCGUCCUUCGGUGGCAGCACGAUGGGAGGCCGCGCCGGUACCCACUUUUCCACUGCGGUAAGCAGAUCACACGUUACGUACGUAGCUCCCAUUUGGCCUAACG ((.....))((((....).))).((..((((((((((((((..((...........))..))))))...........((((.....))))..))))))))..))...... ( -33.00, z-score = 0.60, R) >consensus GAUCGAGGCCUCCUUUGGCGGCUCCUCGCUGGGAUCGCGCAAGGGGACGCACUUCAACACGCAGGUAAGUUCCACAGCCAUUCACUUC_C____________________ ....((((...((((((((((..(.......)..)))).))))))......))))....................................................... (-11.47 = -11.04 + -0.43)
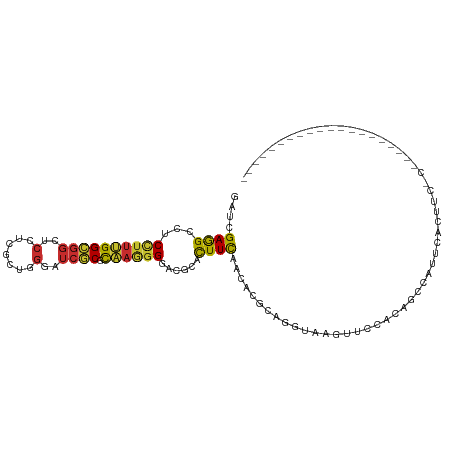
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:55 2011