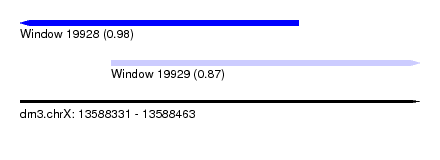
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,588,331 – 13,588,463 |
| Length | 132 |
| Max. P | 0.978678 |

| Location | 13,588,331 – 13,588,423 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Shannon entropy | 0.44824 |
| G+C content | 0.48493 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.92 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
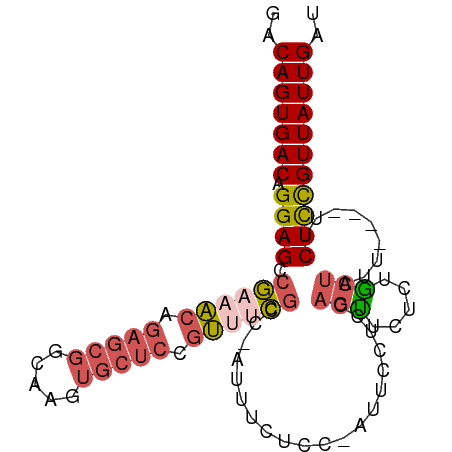
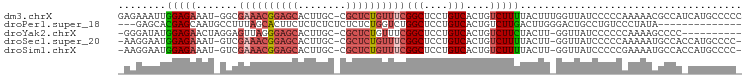
>dm3.chrX 13588331 92 - 22422827 GACAGUGACAGGAGCCGAAACAGAGCGGCAAGUGCUCCGUUUCGCC-AUUUCUCCAAUUUCUCCAGUUCUCUGCUAUU----UUCUCCGUUAUUGAU ..(((((((.((((.((((((.(((((.....))))).))))))..-................(((....))).....----..))))))))))).. ( -26.40, z-score = -2.48, R) >dp4.chrXL_group1e 1228142 92 - 12523060 GACAGUGACAGGAGCCAGACCAGAGAGAGAGAGAGAGAGGAGUGCUAGAGGC----AUUGCUCGUGCUC-GUGCUCGUGCUUUGCUCUGUUAUUGAU ..(((((((((.(((.(((.((..(((...(((...(((.(((((.....))----))).)))...)))-...))).)).))))))))))))))).. ( -36.70, z-score = -2.59, R) >droYak2.chrX 7871819 96 - 21770863 GACAGUGACAGGAGCCGAAACAGAGCGGCAAGUGCUCCCUAACUCCUAGUUCUCC-AUAUCCCCAUUUCUCCACUUUUGCAAUUCUUCGUUAUUGAU ..(((((((((((((((........))))(((((.....................-...............))))).......)))).))))))).. ( -16.22, z-score = 0.17, R) >droSec1.super_20 226991 91 - 1148123 GACAGUGACAGGAGCCGAAACAGAGCGGCAAGUGCUCCGUUUCGAC-AUUUCUCC-AUUCCUUCAGUUCUCUGCUAUU----UUCUCCGUUAUUGAU ..(((((((.((((.((((((.(((((.....))))).))))))..-........-.......(((....))).....----..))))))))))).. ( -26.90, z-score = -2.53, R) >droSim1.chrX 10416415 91 - 17042790 GACAGUGACAGGAGCCGAAACAGAGCGGCAAGUGCUCCGUUUCGAC-AUUUCUCC-AUUCCUUCAGUUCUCUGCUAUU----UUCUCCGUUAUUGAU ..(((((((.((((.((((((.(((((.....))))).))))))..-........-.......(((....))).....----..))))))))))).. ( -26.90, z-score = -2.53, R) >consensus GACAGUGACAGGAGCCGAAACAGAGCGGCAAGUGCUCCGUUUCGCC_AUUUCUCC_AUUCCUCCAGUUCUCUGCUAUU____UUCUCCGUUAUUGAU ..(((((((.((((.((((((.(((((.....))))).))))))....................(((.....))).........))))))))))).. (-17.60 = -18.92 + 1.32)

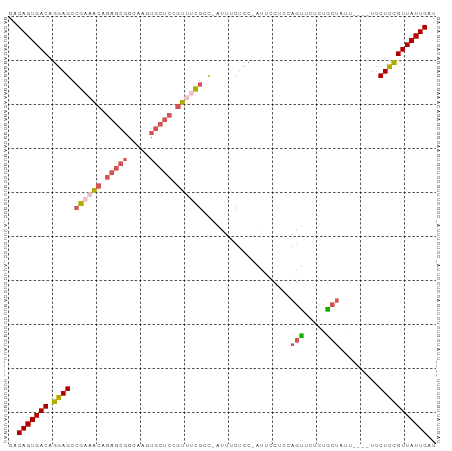
| Location | 13,588,361 – 13,588,463 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.56668 |
| G+C content | 0.51944 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.86 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13588361 102 + 22422827 GAGAAAUUGGAGAAAU-GGCGAAACGGAGCACUUGC-CGCUCUGUUUCGGCUCCUGUCACUGUCUUUUACUUUGGUUAUCCCCCAAAAACGCCAUCAUGCCCCC ........((....((-(((((((((((((......-.))))))))))(((....)))............(((((.......)))))...)))))....))... ( -29.60, z-score = -2.14, R) >droPer1.super_18 76957 87 + 1952607 ---GAGCACGAGCAAUGCCUUUAGCACUUCUCUCUCUCUCUCUGGUCUGGCUCCUGUCACUGUCUUGACUUGGGACUGCCUGUCCCUAUA-------------- ---(((...(((...(((.....)))...)))...))).....((.(.(((((((((((......))))..))))..))).).)).....-------------- ( -20.20, z-score = -0.20, R) >droYak2.chrX 7871853 91 + 21770863 -GGGAUAUGGAGAACUAGGAGUUAGGGAGCACUUGC-CGCUCUGUUUCGGCUCCUGUCACUGUCUUCUACUU-GGUUAUCCCCCCAAAAGCCCC---------- -(((((((.(((...((((((.((((((((......-.))))).....(((....))).))).)))))))))-.).))))))............---------- ( -26.00, z-score = 0.28, R) >droSec1.super_20 227021 99 + 1148123 -AAGGAAUGGAGAAAU-GUCGAAACGGAGCACUUGC-CGCUCUGUUUCGGCUCCUGUCACUGUCUUUUACUU-GGUUAUCCCCCAAAAAUGCCACCAUGCCCC- -..((.((((.((...-(((((((((((((......-.))))))))))))).....)).............(-(((..............)))))))).))..- ( -30.64, z-score = -2.83, R) >droSim1.chrX 10416445 99 + 17042790 -AAGGAAUGGAGAAAU-GUCGAAACGGAGCACUUGC-CGCUCUGUUUCGGCUCCUGUCACUGUCUUUUACUU-GGUUAUCCCCCGAAAAUGCCACCAUGCCCC- -..((.((((.((...-(((((((((((((......-.))))))))))))).....)).............(-(((..((....))....)))))))).))..- ( -30.70, z-score = -2.64, R) >consensus _AAGAAAUGGAGAAAU_GGCGAAACGGAGCACUUGC_CGCUCUGUUUCGGCUCCUGUCACUGUCUUUUACUU_GGUUAUCCCCCAAAAAUGCCA_CAUGCCCC_ ........(((((.......((((((((((........))))))))))(((....)))....)))))..................................... (-13.26 = -14.86 + 1.60)
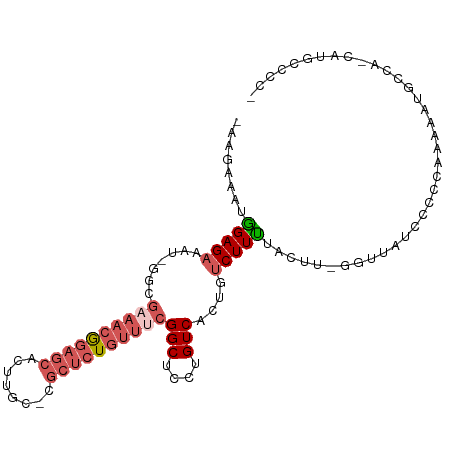
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:52 2011