
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,341,756 – 11,341,864 |
| Length | 108 |
| Max. P | 0.981266 |

| Location | 11,341,756 – 11,341,864 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.02 |
| Shannon entropy | 0.51641 |
| G+C content | 0.51618 |
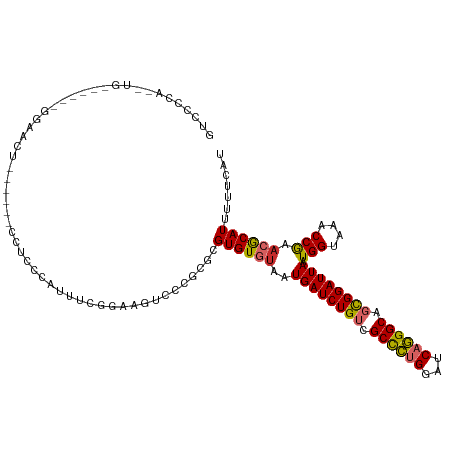
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.59 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
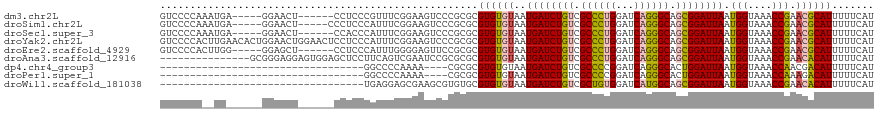
>dm3.chr2L 11341756 108 - 23011544 GUCCCCAAAUGA-----GGAACU------CCUCCCGUUUCGGAAGUCCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU ...((.(((((.-----(((...------..)))))))).)).........((((......((((((((.((((((...)))))).)))))))).(((....))).))))......... ( -35.60, z-score = -1.75, R) >droSim1.chr2L 11147936 109 - 22036055 GUCCCCAAAUGA-----GGAACU-----CCCUCCCAUUUCGGAAGUCCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU ...((.(((((.-----(((...-----...)))))))).)).........((((......((((((((.((((((...)))))).)))))))).(((....))).))))......... ( -36.20, z-score = -2.02, R) >droSec1.super_3 6736321 108 - 7220098 GUCCCCAAAUGA-----GGAACU------CCACCCAUUUCGGAAGUCCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU ........((((-----(((..(------((.........)))..)))...((((......((((((((.((((((...)))))).)))))))).(((....))).)))).....)))) ( -34.70, z-score = -1.64, R) >droYak2.chr2L 7737559 119 - 22324452 GUCCCCACUUGAACACUGGAACUGGAACUCCUCCCAUUUCGGAAGUCCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU .........((((..((((((..(((.....)))..)))))).........((((......((((((((.((((((...)))))).)))))))).(((....))).))))....)))). ( -36.20, z-score = -0.99, R) >droEre2.scaffold_4929 12538964 108 + 26641161 GUCCCCACUUGG-----GGAGCU------CCUCCCAUUUGGGGAGUUCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU ..........(.-----((((((------((((......)))))))))).)((((......((((((((.((((((...)))))).)))))))).(((....))).))))......... ( -46.30, z-score = -3.27, R) >droAna3.scaffold_12916 12055841 104 + 16180835 ---------------GCGGGAGGAGUGGAGCUCCUUCAGUCGAAUCCGCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACACAUUUUUCAU ---------------((((((((((.....))))))).(.((....))).)))((((((..((((((((.((((((...)))))).)))))))).(((....))).))))))....... ( -42.40, z-score = -2.94, R) >dp4.chr4_group3 6779906 81 - 11692001 ----------------------------------GGCCCCAAAA----CGCGCGUGUGUAAUGAUCUGUCGCCCCGGAUCAGGGCACUGGAUUAAUGGUAAACCAACGACAUUUUUCAU ----------------------------------.((((....(----(((....))))..(((((((......)))))))))))....((..((((............))))..)).. ( -21.00, z-score = -0.04, R) >droPer1.super_1 8228926 81 - 10282868 ----------------------------------GGCCCCAAAA----CGCGCGUGUGUAAUGAUCUGUCGCCCCGGAUCAGGGCACUGGAUUAAUGGUAAACCAAAGACAUUUUUCAU ----------------------------------.((((....(----(((....))))..(((((((......)))))))))))....((..((((............))))..)).. ( -21.00, z-score = -0.27, R) >droWil1.scaffold_181038 249773 85 + 637489 ----------------------------------UGAGGAGCGAAGCGUGUGCGUGUGUAAUGAUCUGUCGCUGUGGAUCAUGGCAGCGGAUUAAUGGUAAACCGAACACAUUUUUCAU ----------------------------------(((((((((.......)))((((((..((((((((.((((((...)))))).)))))))).(((....))).)))))))))))). ( -25.70, z-score = -1.18, R) >consensus GUCCCCA__UG______GGAACU______CCUCCCAUUUCGGAAGUCCCGCGCGUGUGUAAUGAUCUGUCGCCCUGGAUCAGGGCAGCGGAUUAAUGGUAAACCGAACGCAUUUUUCAU .....................................................((((((..((((((((.((((((...)))))).)))))))).(((....))).))))))....... (-23.70 = -23.59 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:08 2011