
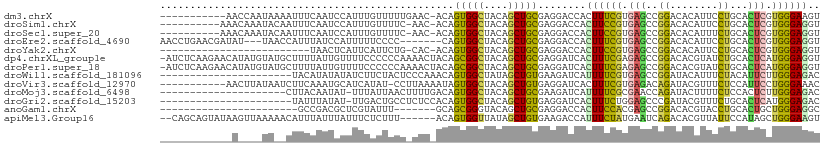
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,582,134 – 13,582,232 |
| Length | 98 |
| Max. P | 0.726189 |

| Location | 13,582,134 – 13,582,232 |
|---|---|
| Length | 98 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 62.14 |
| Shannon entropy | 0.80811 |
| G+C content | 0.46737 |
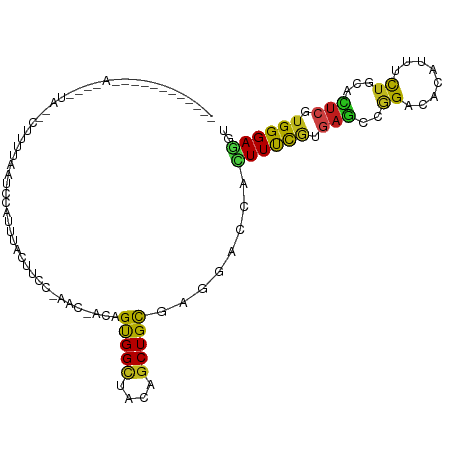
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -10.26 |
| Energy contribution | -9.42 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13582134 98 - 22422827 -----------AACCAAUAAAAUUUCAAUCCAUUUGUUUUUGAAC-ACAGUGGCUACAGCUGCGAGGACCACUUUCGUGAGCCGGACACAUUCCUGCACUCGUGGGAAGU -----------..........(((((...(((((((((....)))-).)))))......(..((((...(((....))).((.(((.....))).)).))))..)))))) ( -24.00, z-score = -0.56, R) >droSim1.chrX 10410744 98 - 17042790 ----------AAACAAAUACAAUUUCAAUCCAUUUGUUUUC-AAC-ACAGUGGCUACAGCUGCGAGGACCACUUCCGUGAGCCGGACACAUUCCUGCACUCGUGGGAGGU ----------...........(((((...((((((((....-.))-).)))))((((((.(((.((((.....((((.....)))).....))))))))).))))))))) ( -22.60, z-score = -0.03, R) >droSec1.super_20 221645 98 - 1148123 ----------AAACAAAUACAAUUUCAAUCCAUUUGUUUUC-AAC-ACAGUGGCUACAGCUGCGAGGACCACUUCCGUGAGCCGGACACAUUUCUGCACUCGUGGGAGGU ----------..................(((...(((....-.))-)..(..((....))..)..)))...((((((((((.((((......))))..)))))))))).. ( -22.80, z-score = -0.25, R) >droEre2.scaffold_4690 5447665 100 + 18748788 AACCUGAACGAUAU---UAACCAUUUAUCCAUUUUUCCCC-------CAGUGGCUACAGCUGCGAGGACCACUUUCGUGAGCCGGACACAUUCCUGCACUCGUGGGAGGU .((((....((((.---........)))).........((-------(((..((....))..)(((...(((....))).((.(((.....))).)).))).)))))))) ( -26.20, z-score = -0.84, R) >droYak2.chrX 7864568 83 - 21770863 -------------------------UAACUCAUUCAUUCUG-CAC-ACAGUGGCUACAGCUGCGAGGACCACUUCCGUGAGCCGGACACAUUCCUGCACUCGUGGGAGGU -------------------------...........(((.(-(.(-((.(..((....))..)..(((.....)))))).)).)))....((((..(....)..)))).. ( -22.60, z-score = 0.12, R) >dp4.chrXL_group1e 1225084 109 - 12523060 -AUCUCAAGAACAUAUGUAUGCUUUUAUUGUUUUCCCCCCAAAACUACAGCGGCUACAGCUGCGAGGAUCACUUUCGAGAGCCGGACACGUAUCUGCACUCAUGGGAGGU -......((((((.(((........))))))))).((((((........(((((....)))))(((((......)).(((..((....))..)))...))).)))).)). ( -25.50, z-score = 0.10, R) >droPer1.super_18 73830 109 - 1952607 -AUCUCAAGAACAUAUGUAUGCUUUUAUUGUUUUCCCCCCAAAACUACAGCGGCUACAGCUGCGAGGAUCACUUUCGAGAGCCGGACACGUAUCUGCACUCAUGGGAGGU -......((((((.(((........))))))))).((((((........(((((....)))))(((((......)).(((..((....))..)))...))).)))).)). ( -25.50, z-score = 0.10, R) >droWil1.scaffold_181096 7637888 88 - 12416693 ----------------------UACAUAUAUAUCUUCUACUCCCAAACAGUGGCUAUAGCUGUGAAGAUCAUUUUCGUGAGCCGGAUACAUUUCUACAUUCUUGGGAGAC ----------------------.................(((((((.....((((...((.(((.....)))....)).))))(((......)))......))))))).. ( -20.00, z-score = -1.34, R) >droVir3.scaffold_12970 29211 98 - 11907090 -----------AACUUAUAAUCUUCAAAUGCAUCAUAU-CCUUAAAAUAGUGGCUACAGCUGUGAGGAUCACUUUCGUGAGACAGAUACGUUUCUCCAUUCCUGGGAAAC -----------........((((((............(-((((...(((((.......))))))))))((((....)))))).))))..(((((.(((....)))))))) ( -19.50, z-score = -0.62, R) >droMoj3.scaffold_6498 388811 88 + 3408170 ---------------------CUUACAAUAU-UUUAUUAACUUUUGACAGUGGCUACAGCUGCGAAGAUCAUUUUCGCGAACCAGAUACUUUUCUCCACUCUUGGGAGAC ---------------------..........-........((((..(.(((((.......((((((((...))))))))....(((......)))))))).)..)))).. ( -19.00, z-score = -2.44, R) >droGri2.scaffold_15203 8930363 87 - 11997470 ----------------------UAUUUAUAU-UUGACUGCCUCUCCACAGUGGCUACAGCUGUGAGGAUCACUUUCUGGAGCCCGAUACGUUUCUGCACUCAUGGGAGAC ----------------------.........-.........(((((.(((.((((.(((..(((.....)))...))).))))).....((....)).....))))))). ( -20.10, z-score = 0.17, R) >anoGam1.chrX 20311919 80 + 22145176 -----------------------GCCGACGCUCGUAUUU-------GCAGCGGGUACAGCUGCGAGGACCACUUCCACGAGCCGGACACGUACCUGCACUGCUGGGAGGC -----------------------(((..(((((((..((-------(((((.......)))))))(((.....)))))))))(((.((.((......))))))))..))) ( -29.90, z-score = -0.77, R) >apiMel3.Group16 4511259 102 - 5631066 --CAGCAGUAUAAGUUAAAAACAUUUAUUUAUUUCUCUUU------ACAGUGGUUAUAGCUGUGAAGACCAUUUCUAUGAAUCAGACACGUUAUUCCAUAGCUGGGAAGU --............................((((((((((------(((((.......))))))))))(((...(((((.................))))).)))))))) ( -18.03, z-score = -0.16, R) >consensus ____________A____UA__CUUUUAAUCCAUUUACUUCC_AAC_ACAGUGGCUACAGCUGCGAGGACCACUUUCGUGAGCCGGACACAUUUCUGCACUCGUGGGAGGU .................................................(((((....)))))........((((((.(((..((........))...))).)))))).. (-10.26 = -9.42 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:49 2011