| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,543,443 – 13,543,562 |
| Length | 119 |
| Max. P | 0.964495 |

| Location | 13,543,443 – 13,543,562 |
|---|---|
| Length | 119 |
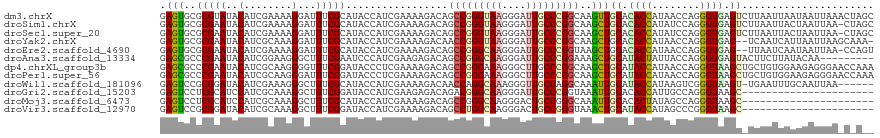
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Shannon entropy | 0.52435 |
| G+C content | 0.50852 |
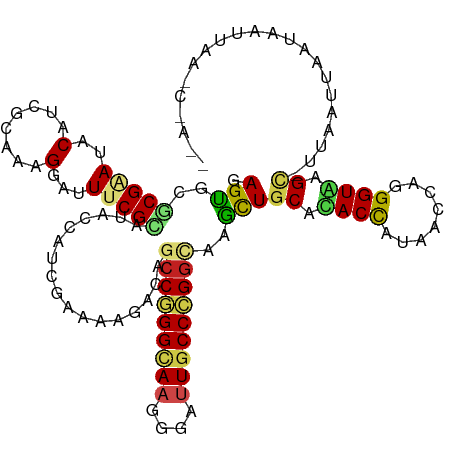
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.12 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13543443 119 + 22422827 GAGUGCGCGUAUACAUCGAAAAGGAUUUCGCAUACCAUCGAAAAGACAGCCGGGUAAGGGAUUGCCCGGCAAGUUGCACACCAUAACCAGGGUGAGUCUUAAUUAAUAAUUAAACUAGC (.(((.(((.....(((......)))..))).))))......(((((.(((((((((....)))))))))........((((........)))).)))))................... ( -33.00, z-score = -2.16, R) >droSim1.chrX 10376649 118 + 17042790 GAGUGCGCGAAUACAUCGAAAAGGAUUUCGCAUACCAUCGAAAAGACAGCCGGGUAAGGGAUUGCCCGGCAAGCUGCACACCAUAUCCAGGGUGAGUCUUAAUUACUAAUUAA-CUAGC .((((.(((((...(((......))))))))...........(((((.(((((((((....)))))))))........((((........)))).)))))...))))......-..... ( -36.00, z-score = -2.74, R) >droSec1.super_20 183753 118 + 1148123 GAGUGCGCGAAUACAUCGAAAAGGAUUUCGCAUACCAUCGAAAAGACAGCCGGGUAAGGGAUUGCCCGGCAAGCUGCACACCAUAUCCAGGGUGAGUCUUAAUUACUAAUUAA-CUAGC .((((.(((((...(((......))))))))...........(((((.(((((((((....)))))))))........((((........)))).)))))...))))......-..... ( -36.00, z-score = -2.74, R) >droYak2.chrX 7826463 116 + 21770863 GAGUGCGCGAAUACAUCGCAAAGGAUUUCGCAUACCAUCGAAAAGACAACCGGGUAAGGGAUUGCCCGGCAAGCUGCACACCAUAACCAGGGUGAG--UCAAUCAUUAAUUAAGCAAA- ...(((((((.....))))....(((((((........))))).(((..((((((((....)))))))).........((((........)))).)--))..)).........)))..- ( -32.10, z-score = -1.96, R) >droEre2.scaffold_4690 5409257 116 - 18748788 GAGUGCGCGAAUACAUCGAAAAGGAUUUCGCAUACCAUCGAAAAGACAGCCGGGCAAGGGAUUGCCCGGUAAGCUGCACACCAUAACCAGGGUGAG--UUAAUCAAUAAUUAA-CCAGU ..(((((((((...(((......))))))))............((...(((((((((....)))))))))...))))))...........(((.((--(((.....))))).)-))... ( -35.40, z-score = -3.02, R) >droAna3.scaffold_13334 459981 110 + 1562580 GAGCGCCCGAAUACAUCGGAAGGGCUUUCGAAUCCCAUCGAAGAGACAGCCGGGCAAGGGAUUGCCCGGAAAGCUGCAUACUAUUACCAGGGUGAGUACUUCUUAUACAA--------- ...(((((.......((....))((((((((......))))))((....((((((((....))))))))....))))............)))))................--------- ( -33.30, z-score = -1.06, R) >dp4.chrXL_group3b 81956 119 + 386183 GAGCGCCCGAAUACAUCGCAAGGGGUUUCGGAUACCCUCGAAAAGACAGCCGGGAAAGGGCUUGCCCGGCAAGCUGCAUACCAUAACCAGGGUAAGCUGCUGUGGAAGAGGGAACCAAA ......(((((.((..(....)..)))))))...(((((.........((((((.((....)).)))))).(((.((.((((........)))).)).)))......)))))....... ( -43.40, z-score = -1.57, R) >droPer1.super_56 103949 119 + 431378 GAGCGCCCGAAUACAUCGCAAGGGAUUUCGGAUACCCUCGAAAAGACAGCCGGGAAAGGGCUUGCCCGGCAAGCUGCAUACCAUAACCAGGGUAAGCUGCUGUGGAAGAGGGAACCAAA ......(((((....((....))...)))))...(((((.........((((((.((....)).)))))).(((.((.((((........)))).)).)))......)))))....... ( -42.70, z-score = -1.86, R) >droWil1.scaffold_181096 896403 112 - 12416693 GAGUCCGCGGAUACAUCGAAAGGGCUUUCGCAUACCAUCGAAAAGACAACCAGGCAAAGGGUUGCCAGGCAAAUUGCAUACCAUAAGUCGGGUAAGU-UGAAUUUGCAAUUAA------ ......(((((....((....))...))))).....................(((((....)))))..(((((((((.((((........)))).))-..)))))))......------ ( -26.90, z-score = -0.53, R) >droGri2.scaffold_15203 9456581 97 + 11997470 GAGUCCUCGCAUCCAUCGCAAAGGCUUUCGGAUACCAUCGAAGAGACAGACGGGCAAGGGAUUGCCCGGUAAAUUGCACACCAUUGCCAGGGUAAGC---------------------- ...((((.(((......((((..((((((((......)))))).......(((((((....)))))))))...)))).......))).)))).....---------------------- ( -26.52, z-score = -0.15, R) >droMoj3.scaffold_6473 11978028 97 - 16943266 GAGUCCUCGCAUCCAUCGCAAAGGCUUUCGCAUACCAUCGAAAAGACAGCCGGGCAAGGGACUGCCGGGCAAAUUGCACACUAUAGCCAGGGUAAGC---------------------- ...((((.((.......))...((((((((........))))).....(((.((((......)))).)))...............))))))).....---------------------- ( -24.70, z-score = 0.52, R) >droVir3.scaffold_12970 6519292 97 - 11907090 GAGUCCGCGGAUACAUCGCAAAGGCUUUCGGAUACCAUCGAAAAGACAGCCUGGCAAGGGACUGCCGGGUAAACUGCAUACCAUAGCCCGGGUAAGC---------------------- .(((((((((.....))))..((((((((((......))))).....)))))......))))).((((((...............))))))......---------------------- ( -29.06, z-score = -0.43, R) >consensus GAGUGCGCGAAUACAUCGCAAAGGAUUUCGCAUACCAUCGAAAAGACAGCCGGGCAAGGGAUUGCCCGGCAAGCUGCACACCAUAACCAGGGUAAGC_UUAAUUAAUAAUUAA_C_A__ .(((..(((((..(........)...))))).................(((((((((....)))))))))..)))((.((((........)))).))...................... (-21.52 = -21.12 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:44 2011