| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,328,738 – 11,328,832 |
| Length | 94 |
| Max. P | 0.955892 |

| Location | 11,328,738 – 11,328,832 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.53596 |
| G+C content | 0.46401 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
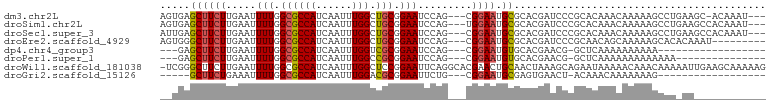
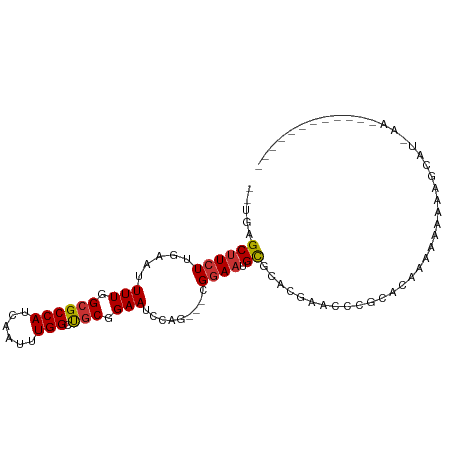
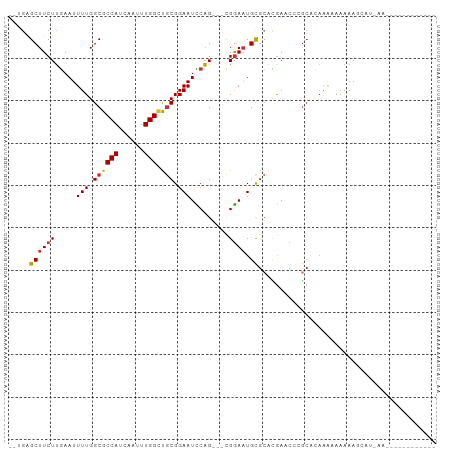
>dm3.chr2L 11328738 94 + 23011544 AGUGAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUGCGGAAUCCAG---CGGAAUGCGCACGAUCCCGCACAAACAAAAAGCCUGAAGC-ACAAAU--- ..((.(((((..(..(((((..((((......))))(((((.(((..(---((.....)))..))).)))))....)))))..)..)))))-.))...--- ( -31.00, z-score = -2.10, R) >droSim1.chr2L 11130385 95 + 22036055 AGUGAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUGCGGAAUCCAG---UGGAAUGCGCACGAUCCCGCACAAACAAAAAGCCUGAAGCCACAAAU--- .(((.(((((..(..(((((..((((......))))(((((.(((..(---((.....)))..))).)))))....)))))..)..))))))))....--- ( -32.60, z-score = -2.72, R) >droSec1.super_3 6723428 95 + 7220098 AUUGAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUGCGGAAUCCAG---CGGAAUGCGCACGAUCCCGCACAAACAAAAAGCCUGAAGCCACAAAU--- ..((.(((((..(..(((((..((((......))))(((((.(((..(---((.....)))..))).)))))....)))))..)..))))))).....--- ( -30.60, z-score = -2.21, R) >droEre2.scaffold_4929 12525033 89 - 26641161 AGUGGGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUGCGGAAUCCAG---CGGAAUGCGCACGAUCCCGCAACAGCAAAAAGCACACAAAU--------- .(((.((((.(((.........((((......))))(((((.(((..(---((.....)))..))).)))))....))).)))).)))....--------- ( -29.70, z-score = -1.64, R) >dp4.chr4_group3 6764470 76 + 11692001 ---GAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGUCGCGGAAUCCAG---CGGAAUGUGCACGAACG-GCUCAAAAAAAAAA------------------ ---(((((..(((...(((.((((((......))).))).)))....(---((.....))).)))..)-))))..........------------------ ( -20.20, z-score = -0.74, R) >droPer1.super_1 8213321 79 + 10282868 ---GAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCCGCGGAAUCCAG---CGGAAUGUGCACGAACG-GCUCAAAAAAAAAAAAA--------------- ---(((((..(((...(((.((((((......))).))).)))....(---((.....))).)))..)-)))).............--------------- ( -20.20, z-score = -0.48, R) >droWil1.scaffold_181038 236568 100 - 637489 -UCGGGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUCCGGAAUUCAGGCACGAACUGCAACUAAAGCAGAAUAAAAACAAACAAAAAUUGAAGCAAAAAG -....((((((((((((((((.((((......)))).))))))))))).......((((.......))))....................)))))...... ( -31.30, z-score = -4.54, R) >droGri2.scaffold_15126 7133965 74 - 8399593 -----GCUUCUGAAAUUUUGGCGCCAUCAAUUUGGACGCGGAAUUCUG---CGGAAUGCGAGUGAACU-ACAAACAAAAAAAG------------------ -----(((((((....(((.((((((......))).))).))).....---))))).)).((....))-..............------------------ ( -18.60, z-score = -1.83, R) >consensus __UGAGCUUCUUGAAUUUUGGCGCCAUCAAUUUGGCUGCGGAAUCCAG___CGGAAUGCGCACGAACCCGCACAAAAAAAAAGCAU_AA____________ .....(((...........)))((((......)))).(((.(.(((......))).).)))........................................ (-12.50 = -12.43 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:07 2011