| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,534,032 – 13,534,098 |
| Length | 66 |
| Max. P | 0.714693 |

| Location | 13,534,032 – 13,534,098 |
|---|---|
| Length | 66 |
| Sequences | 7 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 65.83 |
| Shannon entropy | 0.65503 |
| G+C content | 0.32152 |
| Mean single sequence MFE | -7.79 |
| Consensus MFE | -3.36 |
| Energy contribution | -4.73 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
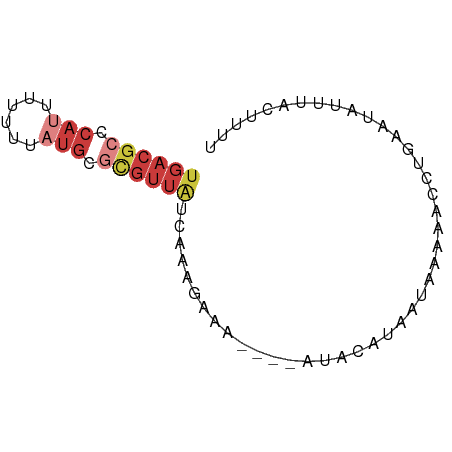
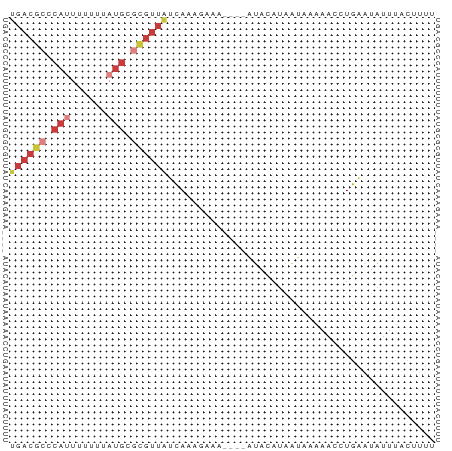
>dm3.chrX 13534032 66 - 22422827 UGACGCCCAUUUUUUUAUGCGCGUUAUCAAAGAAA----AUACAUAAUAAAAACCUGAAUAUUUACUUUU ((((((.(((......))).)))))).........----............................... ( -8.50, z-score = -1.83, R) >droEre2.scaffold_4690 5400095 65 + 18748788 UGACGCCCAUUAUUUUAUGCGCGUUAUCAUCAAAA----AUACAUAAC-AAAACCUGAAUAUUUACUUUU ((((((.(((......))).)))))).........----.........-..................... ( -8.50, z-score = -2.19, R) >droYak2.chrX 7817072 68 - 21770863 --ACGCCCAUUUUUUUAUGCGCGUUAUCAAAGAAAAUACAUACAUAAUAAAAACCUGAAUAUUUACUUUU --((((.(((......))).)))).............................................. ( -5.70, z-score = -0.78, R) >droSec1.super_20 172830 66 - 1148123 UGACGCACAUUUUUUUAUGCGCGUUAUCAAAGAAA----AUACAUAAUAAAAACCUGAAUAUUUACAUUU ((((((.(((......))).)))))).........----............................... ( -9.40, z-score = -1.92, R) >droSim1.chrX 10367350 65 - 17042790 UGACGCCCA-UUUUUUAUGCGCGUUAUCAAAGAAA----AUACAUAAUAAAAACCUGAAUAUUUACUUUU ((((((.((-(.....))).)))))).........----............................... ( -8.30, z-score = -1.77, R) >droPer1.super_56 93370 51 - 431378 UGACGCCCAUCCUUUUCUGC-UGUUGGCA----------GCACUGGACAGACAUUUGUCUUU-------- .........(((.....(((-((....))----------)))..))).((((....))))..-------- ( -11.40, z-score = -0.67, R) >droWil1.scaffold_181089 7107455 53 + 12369635 UGUCACCGAUCUCUUCAGACCC--UACGAAU--------GCAAAAAGCGCAAUGUUUUUUUUU------- .(((...((.....)).)))..--.......--------..(((((((.....)))))))...------- ( -2.70, z-score = 1.39, R) >consensus UGACGCCCAUUUUUUUAUGCGCGUUAUCAAAGAAA____AUACAUAAUAAAAACCUGAAUAUUUACUUUU ((((((.(((......))).))))))............................................ ( -3.36 = -4.73 + 1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:40 2011