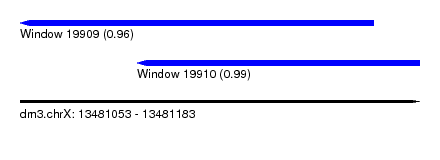
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,481,053 – 13,481,183 |
| Length | 130 |
| Max. P | 0.987966 |

| Location | 13,481,053 – 13,481,168 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.30 |
| Shannon entropy | 0.61667 |
| G+C content | 0.41477 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -8.63 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
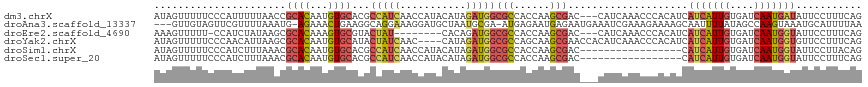
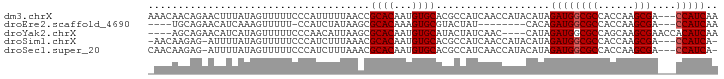
>dm3.chrX 13481053 115 - 22422827 AUAGUUUUUCCCAUUUUUAACCGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC---CAUCAAACCCACAUCAUCAUUGUGAUCAAUGAUAUUCCUUUCAG ...((((...............((((...))))...................((((((((......))).)---)))))))).......(((((((....)))))))........... ( -22.20, z-score = -3.07, R) >droAna3.scaffold_13337 13058491 113 + 23293914 ---GUUGUAGUUCGUUUUAAAUG-AGAAACUGAAGGCAGGAAAGGAUGCUAAUGCGA-AUGAGAAUGAGAAUGAAAUCGAAAGAAAAGCAAUUUUAUAGCCAAGUAAAUGCAUUUUAA ---.((((..((.(((((.....-.))))).))..))))...(((((((...(((..-.((...((((((.((...((....))....)).))))))...)).)))...))))))).. ( -19.30, z-score = -0.56, R) >droEre2.scaffold_4690 5347294 106 + 18748788 AAAGUUUUU-CCAUCUAUAAGCGCACAAAGUGCGUACUAU--------CACAGAUGGCGCCACCAAGCGAC---CAUCAAACCCACAUCAUCAUUGUGAUCAAUGGUAUUCCUUUCAG ((((.....-((((......((((((...)))))).....--------....((((((((......))).)---)))).....((((.......))))....)))).....))))... ( -25.30, z-score = -3.50, R) >droYak2.chrX 7763884 114 - 21770863 AUAGUUUUUCCCAACAUUAAGCGCACAAUGUGCAUACUAUCAAC----CAUAGAUGGCGCCAGCAAGCGAACCACAUCAAACCCACAUCAUCAUUGUGAUCAAUGGUGUUCCUUUCAG ............(((.....((((.....)))).........((----(((.((((.(((......))).....)))).....((((.......))))....))))))))........ ( -20.70, z-score = -0.70, R) >droSim1.chrX 10338868 101 - 17042790 AUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC-----------------CAUCAUUGUGAUCAAUGGUAUUCCUUACAG ...((((...........))))((((...)))).(((((((...........))))))).....(((.((.-----------------.(((((((....)))))))..))))).... ( -22.50, z-score = -2.45, R) >droSec1.super_20 113398 101 - 1148123 AUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC-----------------CAUCAUUGUGAUCAAUGGUAUUCCUUUCAG ...((((...........))))((((...))))..(((((......((((..((((((((......))).)-----------------))))..))))....)))))........... ( -22.00, z-score = -2.32, R) >consensus AUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGAC___CAUCAAACCCACAUCAUCAUUGUGAUCAAUGGUAUUCCUUUCAG ......................((((...))))...(((((...........)))))(((......)))....................(((((((....)))))))........... ( -8.63 = -9.17 + 0.53)
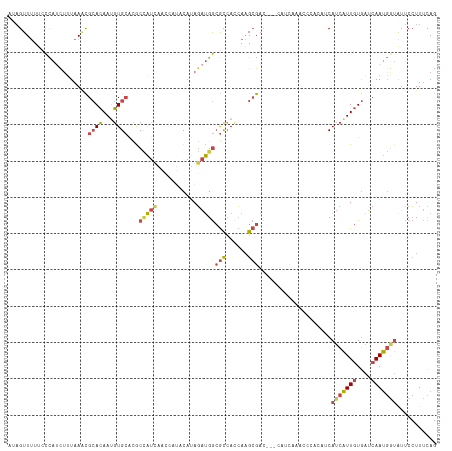
| Location | 13,481,091 – 13,481,183 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Shannon entropy | 0.34859 |
| G+C content | 0.43534 |
| Mean single sequence MFE | -17.24 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
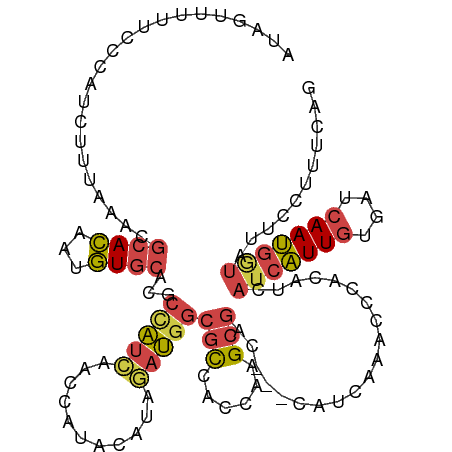
>dm3.chrX 13481091 92 - 22422827 AAACAACAGAACUUUAUAGUUUUUCCCAUUUUUAACCGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGA---CCAUCAA .......((((((....))))))..............((((...))))...................((((((((......))).---))))).. ( -18.00, z-score = -3.58, R) >droEre2.scaffold_4690 5347332 79 + 18748788 ----UGCAGAACAUCAAAGUUUUU-CCAUCUAUAAGCGCACAAAGUGCGUACUAU--------CACAGAUGGCGCCACCAAGCGA---CCAUCAA ----...(((((......))))).-..........((((((...)))))).....--------....((((((((......))).---))))).. ( -20.20, z-score = -3.18, R) >droYak2.chrX 7763922 87 - 21770863 ----AGCAGAACAUCAUAGUUUUUCCCAACAUUAAGCGCACAAUGUGCAUACUAUCAAC----CAUAGAUGGCGCCAGCAAGCGAACCACAUCAA ----.((....((((...(((......))).....((((.....))))...........----....))))((....))..))............ ( -14.10, z-score = -0.78, R) >droSim1.chrX 10338893 89 - 17042790 -AACAAGAG-AUUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGA---CCAUCA- -....((((-((...............))))))...(((.....(((..(((((((...........))))))).)))...))).---......- ( -16.96, z-score = -2.80, R) >droSec1.super_20 113423 90 - 1148123 CAACAAGAG-AUUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGA---CCAUCA- .....((((-((...............))))))...(((.....(((..(((((((...........))))))).)))...))).---......- ( -16.96, z-score = -2.76, R) >consensus _AACAACAGAACUUUAUAGUUUUUCCCAUCUUUAAACGCACAAUGUGCACGCCAUCAACCAUACAUAGAUGGCGCCACCAAGCGA___CCAUCAA .....................................((((...))))...................((((((((......)))....))))).. (-11.64 = -11.84 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:36 2011