
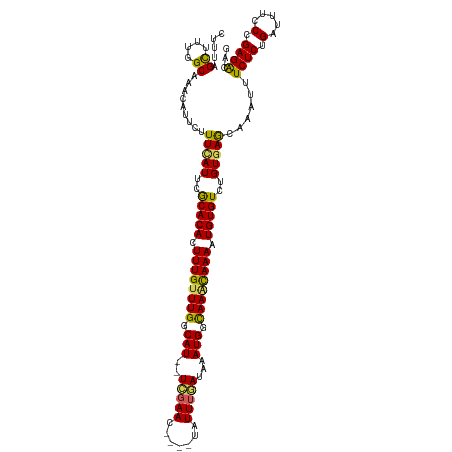
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,465,381 – 13,465,497 |
| Length | 116 |
| Max. P | 0.950404 |

| Location | 13,465,381 – 13,465,495 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.49 |
| Shannon entropy | 0.21144 |
| G+C content | 0.34997 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -23.62 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
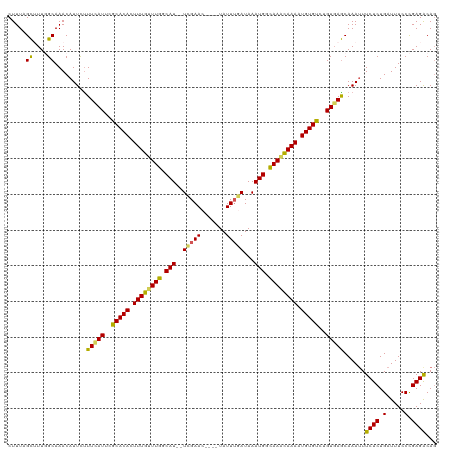
>dm3.chrX 13465381 114 - 22422827 CUUUCGCAUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAU--UCGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAG .....((..((.....)).....((((..(((((.((((((((.(((--((((..----...))))...))).)))))))).)))))..)))))).....((((.(.....).))))... ( -27.10, z-score = -1.78, R) >droSim1.chrX 10324190 114 - 17042790 CUUUUGCUUUGGCAAACAUUCUUUCAUUUGCACACUUUGUUUGGCAU--UCGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAG ..(((((....)))))......(((((..(((((.((((((((.(((--((((..----...))))...))).)))))))).)))))..)))))......((((.(.....).))))... ( -29.50, z-score = -2.61, R) >droSec1.super_20 97994 114 - 1148123 CUUUUGCUUUGGCAAACAUUUUUUCAUUUGCACACUUUGUUUGGCAU--UCGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGGCAG ....((((((((.((((((((.(((((..(((((.((((((((.(((--((((..----...))))...))).)))))))).)))))..))))).)))......)).))).)))))))). ( -33.60, z-score = -3.54, R) >droYak2.chrX 7746652 119 - 21770863 CUUUAGCUUU-GCAAACAUUCCUUUAUUCACACAUUUUGCUUGGCAUUCUCGAACUACAUAUUUAAUAAAUGGUAAAUAAAAUGUGUCUGUUAGCAAAUUUCUCUGAUUUCCUGAGACAG .......(((-((.((((...........((((((((((.(((.((((....................)))).))).)))))))))).)))).)))))..((((.........))))... ( -18.85, z-score = -0.62, R) >droEre2.scaffold_4690 5331450 117 + 18748788 CUUUAGUAUU-GCAAACAUUCUUUUAUUCACACAUUUUGUUUGGCAU--UUGAACUGCAUAUUAAAUAAAUGGCAAGUAAAAUGUGUCUGUGAACAAAAUUCUCUGAUUUCCUGAGACAC ..........-...........(((((..((((((((((((((.(((--(((..............)))))).))))))))))))))..)))))......((((.........))))... ( -22.34, z-score = -0.85, R) >consensus CUUUAGCUUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAU__UCGAAC____UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGACAG .....((....)).........(((((..(((((.((((((((.(((..(((((.......)))))...))).)))))))).)))))..)))))......((((.(.....).))))... (-23.62 = -22.82 + -0.80)

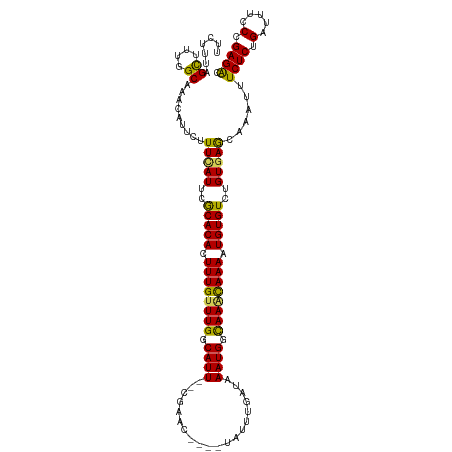
| Location | 13,465,383 – 13,465,497 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.83 |
| Shannon entropy | 0.20543 |
| G+C content | 0.34131 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
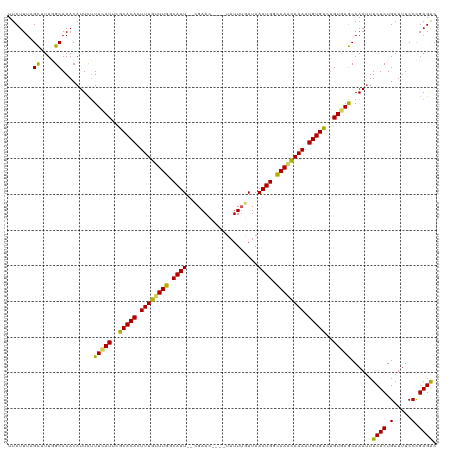
>dm3.chrX 13465383 114 - 22422827 UUCUUUCGCAUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU--CGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGAC .......((..((.....)).....((((..(((((.((((((((.((((--(((..----...))))...))).)))))))).)))))..)))))).....((((.(.....).)))). ( -27.10, z-score = -1.97, R) >droSim1.chrX 10324192 114 - 17042790 UUCUUUUGCUUUGGCAAACAUUCUUUCAUUUGCACACUUUGUUUGGCAUU--CGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGAC ....(((((....)))))......(((((..(((((.((((((((.((((--(((..----...))))...))).)))))))).)))))..)))))......((((.(.....).)))). ( -29.50, z-score = -2.88, R) >droSec1.super_20 97996 114 - 1148123 UUCUUUUGCUUUGGCAAACAUUUUUUCAUUUGCACACUUUGUUUGGCAUU--CGAAC----UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGGC .......(((((((.((((((((.(((((..(((((.((((((((.((((--(((..----...))))...))).)))))))).)))))..))))).)))......)).))).))))))) ( -31.00, z-score = -2.91, R) >droYak2.chrX 7746654 119 - 21770863 UUCUUUAGCUUU-GCAAACAUUCCUUUAUUCACACAUUUUGCUUGGCAUUCUCGAACUACAUAUUUAAUAAAUGGUAAAUAAAAUGUGUCUGUUAGCAAAUUUCUCUGAUUUCCUGAGAC .........(((-((.((((...........((((((((((.(((.((((....................)))).))).)))))))))).)))).)))))..((((.........)))). ( -18.85, z-score = -0.87, R) >droEre2.scaffold_4690 5331452 117 + 18748788 UUCUUUAGUAUU-GCAAACAUUCUUUUAUUCACACAUUUUGUUUGGCAUU--UGAACUGCAUAUUAAAUAAAUGGCAAGUAAAAUGUGUCUGUGAACAAAAUUCUCUGAUUUCCUGAGAC ............-...........(((((..((((((((((((((.((((--((..............)))))).))))))))))))))..)))))......((((.........)))). ( -22.34, z-score = -0.95, R) >consensus UUCUUUAGCUUUGGCAAACAUUCUUUCAUUCGCACACUUUGUUUGGCAUU__CGAAC____UAUUUGAUAAAUGGCAAACAAAAUGUGUCUGUGAGCAAAUUUCUCUGAUUUCCCGAGAC .......((....)).........(((((..(((((.((((((((.((((..((((.......))))...)))).)))))))).)))))..)))))......((((.(.....).)))). (-23.08 = -22.28 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:30 2011