
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,432,676 – 13,432,743 |
| Length | 67 |
| Max. P | 0.991490 |

| Location | 13,432,676 – 13,432,743 |
|---|---|
| Length | 67 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 73.36 |
| Shannon entropy | 0.46089 |
| G+C content | 0.38333 |
| Mean single sequence MFE | -14.69 |
| Consensus MFE | -8.61 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
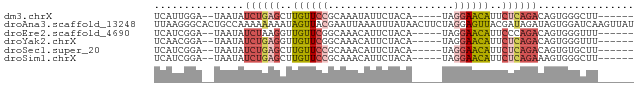
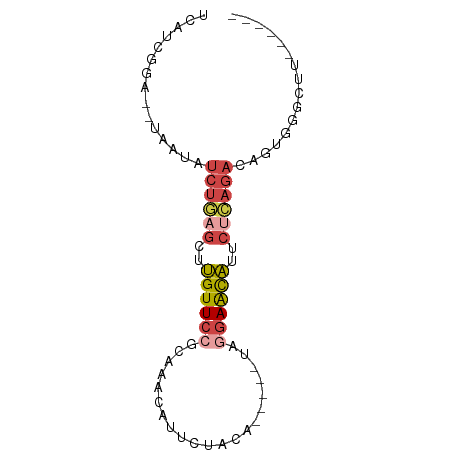
>dm3.chrX 13432676 67 + 22422827 UCAUUGGA--UAAUAUCUGAGCUUGUUCCGCAAAUAUUCUACA-----UAGGAACAUUCUCAGACAGUGGGCUU------ ((((((..--.....((((((..((((((..............-----..))))))..))))))))))))....------ ( -19.30, z-score = -3.07, R) >droAna3.scaffold_13248 2409069 80 + 4840945 UUAAGGGCACUGCCAAAAAAAAUAGUUACGAAUUAAAUUUAUAACUUCUAGGAGUUACGAUAGAUAGUGGAUCAAGUUAU ....((.(((((.(........((((.....))))...(..((((((....))))))..)..).)))))..))....... ( -8.50, z-score = 0.65, R) >droEre2.scaffold_4690 5299744 67 - 18748788 UCAUCGGA--UAAUAUCUAAGGUUGUUCGGCAAACAUUCUACA-----UAGGAACAUUCCCAGACAGUGGGUUU------ ...(((((--((((.......)))))))))......((((...-----..))))....((((.....))))...------ ( -11.00, z-score = -0.09, R) >droYak2.chrX 7713743 67 + 21770863 UCAACGGA--UAAUAUCUGAGGUUGUUCGGCAAACAUUCUACA-----UAGGAACAUUCUCAGACAGUGGGUUU------ ........--.....((((((..(((((...............-----...)))))..))))))..........------ ( -13.37, z-score = -1.20, R) >droSec1.super_20 65829 67 + 1148123 UCAUCGGA--UAAUAUCUGAGCUUGUUCCGCAAACAUUCUACA-----UAGGAACAUUCUCAGACAGUGUGCUU------ .....(.(--((...((((((..((((((..............-----..))))))..))))))...))).)..------ ( -18.99, z-score = -3.17, R) >droSim1.chrX 10309493 67 + 17042790 UCAUCGGA--UAAUAUCUGAGCUUGUUCCGCAAACAUUCUACA-----UAGGAACAUUCUCAGAAAGUGGGCUU------ ........--.....((((((..((((((..............-----..))))))..))))))..........------ ( -16.99, z-score = -2.39, R) >consensus UCAUCGGA__UAAUAUCUGAGCUUGUUCCGCAAACAUUCUACA_____UAGGAACAUUCUCAGACAGUGGGCUU______ ...............((((((..((((((.....................))))))..))))))................ ( -8.61 = -9.50 + 0.89)
| Location | 13,432,676 – 13,432,743 |
|---|---|
| Length | 67 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 73.36 |
| Shannon entropy | 0.46089 |
| G+C content | 0.38333 |
| Mean single sequence MFE | -15.22 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.85 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
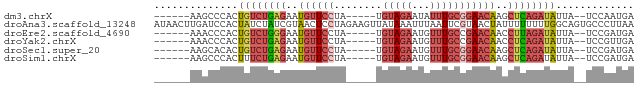
>dm3.chrX 13432676 67 - 22422827 ------AAGCCCACUGUCUGAGAAUGUUCCUA-----UGUAGAAUAUUUGCGGAACAAGCUCAGAUAUUA--UCCAAUGA ------........((((((((..((((((..-----.(((((...)))))))))))..))))))))...--........ ( -19.90, z-score = -3.89, R) >droAna3.scaffold_13248 2409069 80 - 4840945 AUAACUUGAUCCACUAUCUAUCGUAACUCCUAGAAGUUAUAAAUUUAAUUCGUAACUAUUUUUUUUGGCAGUGCCCUUAA ...........((((.((((..........))))(((((..(((...)))..)))))............))))....... ( -6.10, z-score = 0.73, R) >droEre2.scaffold_4690 5299744 67 + 18748788 ------AAACCCACUGUCUGGGAAUGUUCCUA-----UGUAGAAUGUUUGCCGAACAACCUUAGAUAUUA--UCCGAUGA ------........((((((((..(((((...-----.(((((...))))).)))))..))))))))...--........ ( -12.40, z-score = -1.14, R) >droYak2.chrX 7713743 67 - 21770863 ------AAACCCACUGUCUGAGAAUGUUCCUA-----UGUAGAAUGUUUGCCGAACAACCUCAGAUAUUA--UCCGUUGA ------........((((((((..(((((...-----.(((((...))))).)))))..))))))))...--........ ( -15.40, z-score = -3.37, R) >droSec1.super_20 65829 67 - 1148123 ------AAGCACACUGUCUGAGAAUGUUCCUA-----UGUAGAAUGUUUGCGGAACAAGCUCAGAUAUUA--UCCGAUGA ------........((((((((..((((((..-----.(((((...)))))))))))..))))))))...--........ ( -19.90, z-score = -2.92, R) >droSim1.chrX 10309493 67 - 17042790 ------AAGCCCACUUUCUGAGAAUGUUCCUA-----UGUAGAAUGUUUGCGGAACAAGCUCAGAUAUUA--UCCGAUGA ------..........((((((..((((((..-----.(((((...)))))))))))..)))))).....--........ ( -17.60, z-score = -2.72, R) >consensus ______AAACCCACUGUCUGAGAAUGUUCCUA_____UGUAGAAUGUUUGCGGAACAAGCUCAGAUAUUA__UCCGAUGA ..............((((((((..((((((.....................))))))..))))))))............. (-10.01 = -10.85 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:25 2011