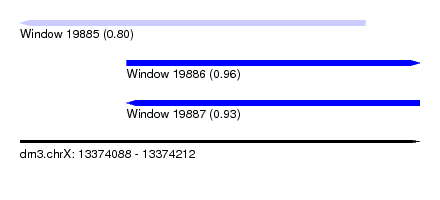
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,374,088 – 13,374,212 |
| Length | 124 |
| Max. P | 0.958320 |

| Location | 13,374,088 – 13,374,195 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Shannon entropy | 0.29539 |
| G+C content | 0.34758 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13374088 107 - 22422827 ------UUUUAAUGCCCUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGG------ ------.......((...(((.(((((((.((((((.....))))))))))).)).........(((((..((((((((......)))))))).))))).)))..))......------ ( -22.90, z-score = -1.25, R) >droAna3.scaffold_13248 2344242 98 - 4840945 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUU---UUCGG------------ ------......(((....)))..(((((.((((((.....)))))))))))((((((.......(((((.....(((((......))))))))))..)---)))))------------ ( -22.60, z-score = -1.96, R) >droEre2.scaffold_4690 5244180 107 + 18748788 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGG------ ------.......((...(((.(((((((.((((((.....))))))))))).)).........(((((..((((((((......)))))))).))))).)))..))......------ ( -22.90, z-score = -1.30, R) >droYak2.chrX 7648232 107 - 21770863 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGC------ ------.......((...(((.(((((((.((((((.....))))))))))).)).........(((((..((((((((......)))))))).))))).)))..))......------ ( -22.90, z-score = -1.19, R) >droSec1.super_20 7565 107 - 1148123 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGG------ ------.......((...(((.(((((((.((((((.....))))))))))).)).........(((((..((((((((......)))))))).))))).)))..))......------ ( -22.90, z-score = -1.30, R) >droSim1.chrX 10263349 107 - 17042790 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGG------ ------.......((...(((.(((((((.((((((.....))))))))))).)).........(((((..((((((((......)))))))).))))).)))..))......------ ( -22.90, z-score = -1.30, R) >droPer1.super_17 1369591 113 + 1930428 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGGUUCUGUUGGGUUUUCGG ------.......((((....(((.....(((((((.....)))))))..(((((((........(((((.....(((((......)))))))))).))))))).))).))))...... ( -26.90, z-score = -1.66, R) >dp4.chrXL_group1e 8899740 113 + 12523060 ------UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGGUUCUGUUGGGUUUUCGG ------.......((((....(((.....(((((((.....)))))))..(((((((........(((((.....(((((......)))))))))).))))))).))).))))...... ( -26.90, z-score = -1.66, R) >droWil1.scaffold_180702 3284749 97 - 4511350 -------UUUAAUACCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA-UUUUUAUGCACUUACACUUACAUAUGUAUGUACAUA-------------- -------............((((((.((((((((((.....)))))))............((((((..-..))))))...............))).))))))...-------------- ( -19.30, z-score = -2.06, R) >droVir3.scaffold_12970 1079696 100 + 11907090 ------UUUAAAUGCCUUGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA--UUUUAUGCAC-UACAUGUGUAAAUGUGU-CUCCUCCGUCU--------- ------............(((.(((((((.((((((.....))))))))))).))..........(((--((...(((((-.....))))))))))..-....)))....--------- ( -22.80, z-score = -2.56, R) >droMoj3.scaffold_6473 10556805 110 - 16943266 AAACCAUUUAAAUGCCUGGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA--UUUUAUGCAC-UACAUGUGUAAAUGUGUGCACGUCCACCUCGG------ ..............((.(((..(((((((.((((((.....))))))))))).)).............--.....(((((-.((((......)))))))))......))).))------ ( -27.40, z-score = -1.69, R) >droGri2.scaffold_15203 9176549 89 + 11997470 ------UUUAAAUGCCCGGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA--UUUUAUGCAC-UACAUGUGUAAAUGUGU--------------------- ------..((((((........)))))).(((((((.....))))))).................(((--((...(((((-.....))))))))))..--------------------- ( -20.60, z-score = -1.25, R) >consensus ______UUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCACUUACAUGUGUAAAUGUGUUCGCUUCGCUUUUGG______ .....................(((((((((((((((.....)))))))............((((((.....))))))...........))))))))....................... (-18.16 = -18.25 + 0.09)

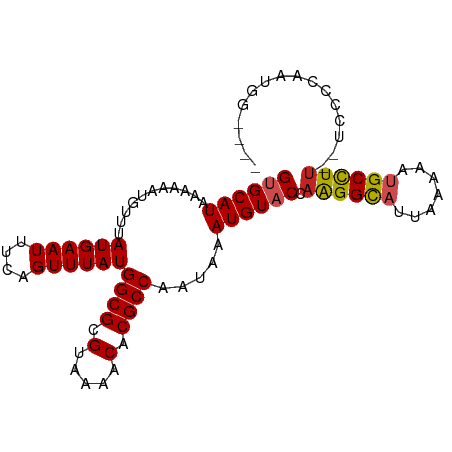
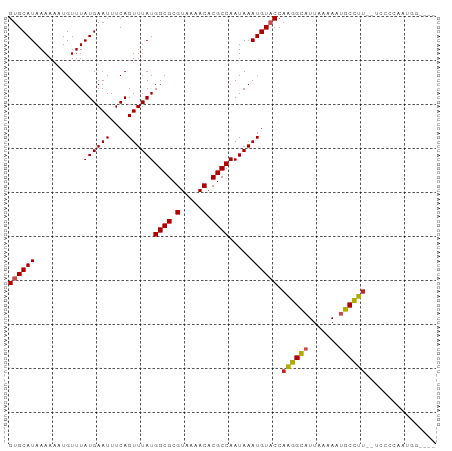
| Location | 13,374,121 – 13,374,212 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Shannon entropy | 0.27994 |
| G+C content | 0.36461 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13374121 91 + 22422827 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAGGGCAUUAAAAAUGCCUU--UCCCCAAUGG---- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))--..........---- ( -21.40, z-score = -1.52, R) >droAna3.scaffold_13248 2344266 83 + 4840945 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUUUG-------------- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))..-------------- ( -21.70, z-score = -2.85, R) >droEre2.scaffold_4690 5244213 91 - 18748788 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUU--UCCCCAAUGG---- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))--..........---- ( -21.70, z-score = -2.18, R) >droYak2.chrX 7648265 91 + 21770863 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUU--UCCCCAAUGG---- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))--..........---- ( -21.70, z-score = -2.18, R) >droSec1.super_20 7598 91 + 1148123 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUU--UCCCCAAUGG---- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))--..........---- ( -21.70, z-score = -2.18, R) >droSim1.chrX 10263382 91 + 17042790 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUU--UCCCCAAUGG---- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((((.....)))))))--..........---- ( -21.70, z-score = -2.18, R) >droPer1.super_17 1369630 96 - 1930428 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAACGCCUUUCUCCUCGCUACUCG- ((((((...........((((((....))))))((((.(.....).)))).....)))))).(((((.........)))))...............- ( -18.90, z-score = -1.65, R) >dp4.chrXL_group1e 8899779 96 - 12523060 GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAACGCCUUGCUCCUCGCUACUCG- ((((((...........((((((....))))))((((.(.....).)))).....))))))((((((.........))))))..............- ( -21.80, z-score = -2.07, R) >droWil1.scaffold_180702 3284774 95 + 4511350 GUGCAUAAAAA-UGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGUAUUAAAA-UGCCUUUUUUUUCCUUCACCAA ((((((.....-.....((((((....))))))((((.(.....).)))).....)))))).(((((((....)-))))))................ ( -19.70, z-score = -2.00, R) >droVir3.scaffold_12970 1079724 89 - 11907090 GUGCAUAAAA--UGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUUCCAAGGCAUUUAAA-UGCUUUUGCUUUGGCAG----- ..((((.(((--(((((..((((....((((((((((.(.....).)))).))))))))))..))))))))..)-)))...(((....))).----- ( -21.80, z-score = -1.21, R) >droGri2.scaffold_15203 9176566 89 - 11997470 GUGCAUAAAA--UGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUUCCCGGGCAUUUAAA-UGCUUUUGCUUUGCCAG----- ..((((.(((--(((((..((((....((((((((((.(.....).)))).))))))))))..))))))))..)-)))..............----- ( -20.10, z-score = -0.59, R) >consensus GUGCAUAAAAAAUGUUUAUGAAUUUCAGUUUAUGGCGCGUAAAACACGCCAAUAAAUGUACCAAGGCAUUAAAAAUGCCUU__UCCCCAAUGG____ ((((((...........((((((....))))))((((.(.....).)))).....)))))).((((((.......))))))................ (-18.31 = -18.38 + 0.08)
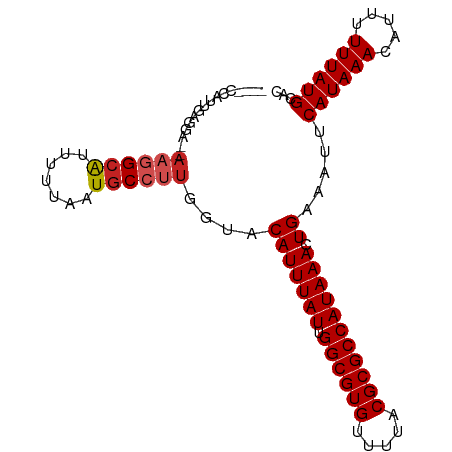
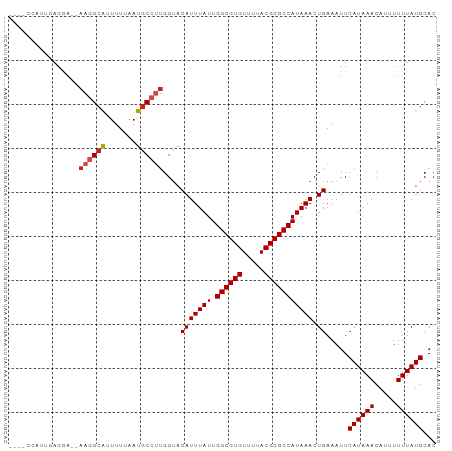
| Location | 13,374,121 – 13,374,212 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Shannon entropy | 0.27994 |
| G+C content | 0.36461 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13374121 91 - 22422827 ----CCAUUGGGGA--AAGGCAUUUUUAAUGCCCUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ----..((..(((.--...............)))..)).(((((((.((((((.....))))))))))).)).....((((((.....))))))... ( -20.49, z-score = -0.92, R) >droAna3.scaffold_13248 2344266 83 - 4840945 --------------CAAAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC --------------..(((((((.....)))))))....(((((((.((((((.....))))))))))).)).....((((((.....))))))... ( -22.30, z-score = -3.29, R) >droEre2.scaffold_4690 5244213 91 + 18748788 ----CCAUUGGGGA--AAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ----...(..(...--(((((((.....)))))))......(((((.((((((.....))))))))))))..)....((((((.....))))))... ( -22.90, z-score = -2.05, R) >droYak2.chrX 7648265 91 - 21770863 ----CCAUUGGGGA--AAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ----...(..(...--(((((((.....)))))))......(((((.((((((.....))))))))))))..)....((((((.....))))))... ( -22.90, z-score = -2.05, R) >droSec1.super_20 7598 91 - 1148123 ----CCAUUGGGGA--AAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ----...(..(...--(((((((.....)))))))......(((((.((((((.....))))))))))))..)....((((((.....))))))... ( -22.90, z-score = -2.05, R) >droSim1.chrX 10263382 91 - 17042790 ----CCAUUGGGGA--AAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ----...(..(...--(((((((.....)))))))......(((((.((((((.....))))))))))))..)....((((((.....))))))... ( -22.90, z-score = -2.05, R) >droPer1.super_17 1369630 96 + 1930428 -CGAGUAGCGAGGAGAAAGGCGUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC -......(((..(((((((((((.....)))))))....(((((((.((((((.....))))))))))).))..............))))..))).. ( -25.30, z-score = -2.12, R) >dp4.chrXL_group1e 8899779 96 + 12523060 -CGAGUAGCGAGGAGCAAGGCGUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC -...(((..((((((((((((((.....))))))))...(((((((.((((((.....))))))))))).)).............)))))).))).. ( -26.90, z-score = -2.37, R) >droWil1.scaffold_180702 3284774 95 - 4511350 UUGGUGAAGGAAAAAAAAGGCA-UUUUAAUACCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA-UUUUUAUGCAC ...(((...(((((..((((.(-(....)).))))....(((((((.((((((.....))))))))))).)).............-)))))...))) ( -18.10, z-score = -0.78, R) >droVir3.scaffold_12970 1079724 89 + 11907090 -----CUGCCAAAGCAAAAGCA-UUUAAAUGCCUUGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA--UUUUAUGCAC -----.(((....)))...(((-(..(((((...((((.(((((((.((((((.....))))))))))).))...))))....))--))).)))).. ( -22.10, z-score = -2.22, R) >droGri2.scaffold_15203 9176566 89 + 11997470 -----CUGGCAAAGCAAAAGCA-UUUAAAUGCCCGGGAACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACA--UUUUAUGCAC -----..............(((-(..(((((.....((((((((((.((((((.....))))))))))).))...))).....))--))).)))).. ( -20.20, z-score = -1.15, R) >consensus ____CCAUUGAGGA__AAGGCAUUUUUAAUGCCUUGGUACAUUUAUUGGCGUGUUUUACGCGCCAUAAACUGAAAUUCAUAAACAUUUUUUAUGCAC ................((((((.......))))))....(((((((.((((((.....))))))))))).)).....((((((.....))))))... (-18.23 = -18.63 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:17 2011