| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,309,228 – 11,309,328 |
| Length | 100 |
| Max. P | 0.549499 |

| Location | 11,309,228 – 11,309,328 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.51499 |
| G+C content | 0.38035 |
| Mean single sequence MFE | -18.51 |
| Consensus MFE | -6.84 |
| Energy contribution | -6.97 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11309228 100 - 23011544 AAAUCGGCCAUACAAAAAAA-------UAAAACCUCCAGCGACA-GCCAUCAAAAUGUAGAUCGUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUCCGAUGGCAAUAA ......(((((.(.......-------.........(((.((((-((.(((........))).))).---))).))).(--------(((.-....)))).........))))))..... ( -19.60, z-score = -1.85, R) >droSim1.chr2L 11113000 107 - 22036055 AAAUCGGCCAUACAAAAAAAAAUAAAAUAAAACCUCUGGCGACA-GCCAUCAAAAUGUAGAUCGUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUCCGAUGGCAAUAA ......((((..........................))))....-((((((..((((..((....))---..))))(.(--------(((.-....)))))........))))))..... ( -21.37, z-score = -1.76, R) >droSec1.super_3 6706260 106 - 7220098 AAAUCGGCCAUACAAAAAAAAA-AAAAUAAAACCUCUGGCGACA-GCCAUCAAAAUGUAGAUCGUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUCCGAUGGCAAUAA ......((((............-.............))))....-((((((..((((..((....))---..))))(.(--------(((.-....)))))........))))))..... ( -21.41, z-score = -1.82, R) >droYak2.chr2L 7706194 107 - 22324452 AAAUCGGCCAUACAAAUAUAUAAAACAAAAAAACUCUGCCGACA-GCCAUCAAAAUGUAGAUCGUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUCCGAUGGCAAUAA ...(((((.............................)))))..-((((((..((((..((....))---..))))(.(--------(((.-....)))))........))))))..... ( -23.05, z-score = -2.76, R) >droEre2.scaffold_4929 12507480 94 + 26641161 AAAUCGGCCAUACAAAAAAA-------------CUCUGCCGACA-GCCAUCAAAAUGUAGAUCGUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUCCGAUGGCAAUAA ...(((((............-------------....)))))..-((((((..((((..((....))---..))))(.(--------(((.-....)))))........))))))..... ( -23.69, z-score = -3.02, R) >droAna3.scaffold_12916 12029532 76 + 16180835 -------------------------------AAAACUGGCGACA-GCCAUCAAAAUGUAGAUCAUUC---GUCAUUGGG--------GGUA-CGAAACCCCAUAAUUUCGAUGGCAAUAA -------------------------------.....((....))-((((((..((((..((....))---..)))).((--------(((.-....)))))........))))))..... ( -22.70, z-score = -2.85, R) >dp4.chr4_group3 6745971 84 - 11692001 --------CAAUCAACAAAA----------------AGUCGACAGCCCAUCAAAAUGUAGAUCAUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUUCGAUGGCAAUAA --------............----------------.(((.(((...........))).))).....---(((((((((--------(((.-....)))).......))))))))..... ( -14.31, z-score = -1.38, R) >droPer1.super_1 8195074 84 - 10282868 --------CAAUCAACAAAA----------------AGUCGACAGCCCAUCAAAAUGUAGAUCAUUC---GUCAUUGAG--------GGUA-CAAAACCCCAUAAUUUCGAUGGCAAUAA --------............----------------.(((.(((...........))).))).....---(((((((((--------(((.-....)))).......))))))))..... ( -14.31, z-score = -1.38, R) >droWil1.scaffold_181038 220868 100 + 637489 AAAUCGAUAAAAAAAAAAA----------------CAGCAGACAGGCCAUCAAAAUGUAGAUCAUUC---GUCAUUGAGGGAAGGAAGGCA-CAAAACCCCAUAAUUUCGAUGGCAAUAA ...................----------------..........((((((..((((..((....))---..))))..(((..........-......)))........))))))..... ( -15.99, z-score = -0.61, R) >droVir3.scaffold_12963 17998324 93 + 20206255 AAAUCGACAACAAAACCGAA---------------AAAAAGGCAACACAUCAAAAUGUAGAUCAUAC---GUCAUUGAA--------GGUAUC-AAAGCCCAUAGUUUCGAUGGCAAUAA ..(((...............---------------.....(....)((((....)))).))).....---(((((((((--------(.(((.-.......))).))))))))))..... ( -15.50, z-score = -1.70, R) >droMoj3.scaffold_6500 4944800 93 + 32352404 AAAUCGACAAAUAAACCG-A---------------AAAAAGGCAACACAUCAAAAUGUAGAUCAUAC---GUCAUUGAA--------GCUACCAAAAGCCCAUAGUUUCGAUGGCAAUAA ..(((.............-.---------------.....(....)((((....)))).))).....---(((((((((--------((((...........)))))))))))))..... ( -17.60, z-score = -2.90, R) >droGri2.scaffold_15126 7114556 94 + 8399593 -AAACACCCA-ACAACAACA----------------AAAAAGCAACACAUCAAAAUGUAGAUCAUACUAUGUCACUGAA--------GGUAUCCAAAGCCCAUCGUUUUGAUGGCAAUAA -.........-.........----------------...........((((((((((..(((........)))......--------(((.......)))...))))))))))....... ( -12.60, z-score = -0.50, R) >consensus AAAUCGGCCAUACAAAAAAA_______________CAGGCGACA_GCCAUCAAAAUGUAGAUCAUUC___GUCAUUGAG________GGUA_CAAAACCCCAUAAUUUCGAUGGCAAUAA ......................................................................(((((((..........(((......))).........)))))))..... ( -6.84 = -6.97 + 0.13)
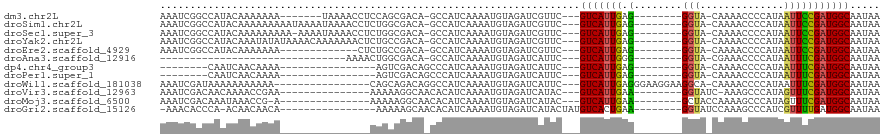


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:04 2011